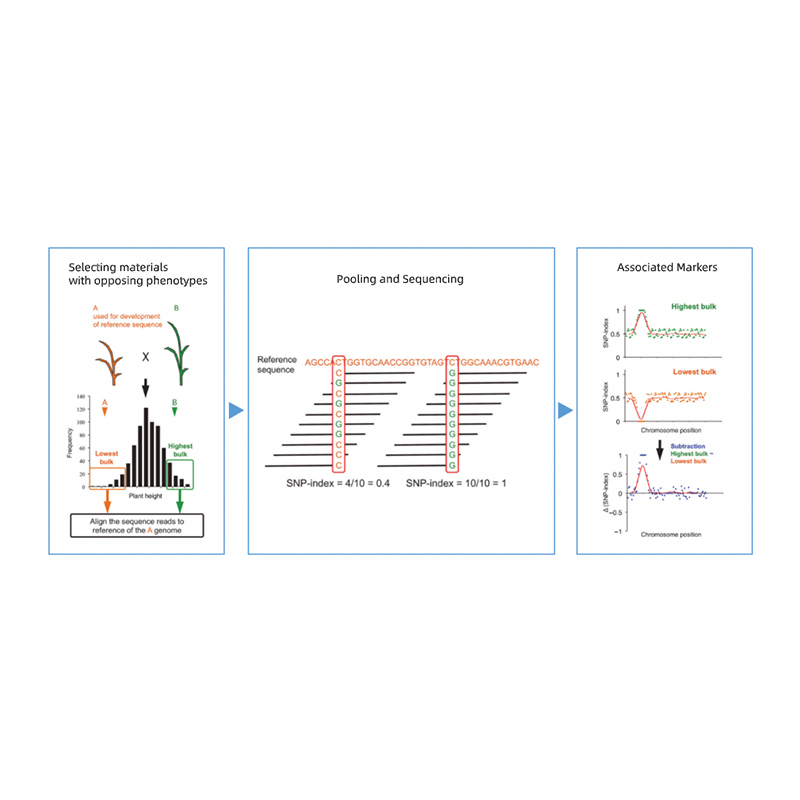
Ukuhlaziywa kwe-Bulked Secregant
Izinzuzo zensizakalo
Tatagi et al., Iphephabhuku lezitshalo, 2013
● Ukuhlelwa kwendawo okunembile: ukuxuba ama-bulks nabangu-30 kuya ku-200 abantu abangu-200 ukuze banciphise umsindo wangemuva; ukubikezela okungafani nokufundwayo okusekelwe endabeni.
● Ukuhlaziywa okuphelele: Isichasiselo se-gene yofuzo elijulile, kufaka phakathi i-NR, SwissProt, Go, Kegg, COG, KOG, njll.
● Isikhathi esiguqukayo ngokushesha: I-Rapid gene yasendaweni ukuzokwenza ngaphakathi kwezinsuku ezingama-45 zomsebenzi.
● Umuzwa obanzi: I-BMK iba negalelo ezinkulungwaneni zezici zasendaweni, emboza izinhlobo ezahlukahlukene njengezitshalo, imikhiqizo yasemanzini, ihlathi, izimbali, izithelo, njll.
Ukucaciswa Kwensiza
Inani labantu:
Ukuhlukaniswa inzalo yabazali abane-phenotypes ephikisayo.
I-EG F2 Progeny, i-Backcrossing (BC), I-Recombivent InBred Line (RIL)
Ukuxuba Ichibi
Ngezimpawu ezifanelekile: abantu abangama-30 kuye kwangama-50 (ubuncane be-20) / inqwaba
Nge-Tratis yenani: I-5% ephezulu kuya ku-10% abantu abane-phenotypes ebabazekayo kubo bonke abantu (ubuncane 30 + 30).
Ukujula okuncomekayo kokulandelana
Okungenani i-20x / umzali no-1x / inzalo / inzalo echibini lokuxuba inzalo elingu-30 + 30 umuntu ngamunye, ukujula okubuyiselwe kuzoba ngu-30x ngobuningi ngakunye)
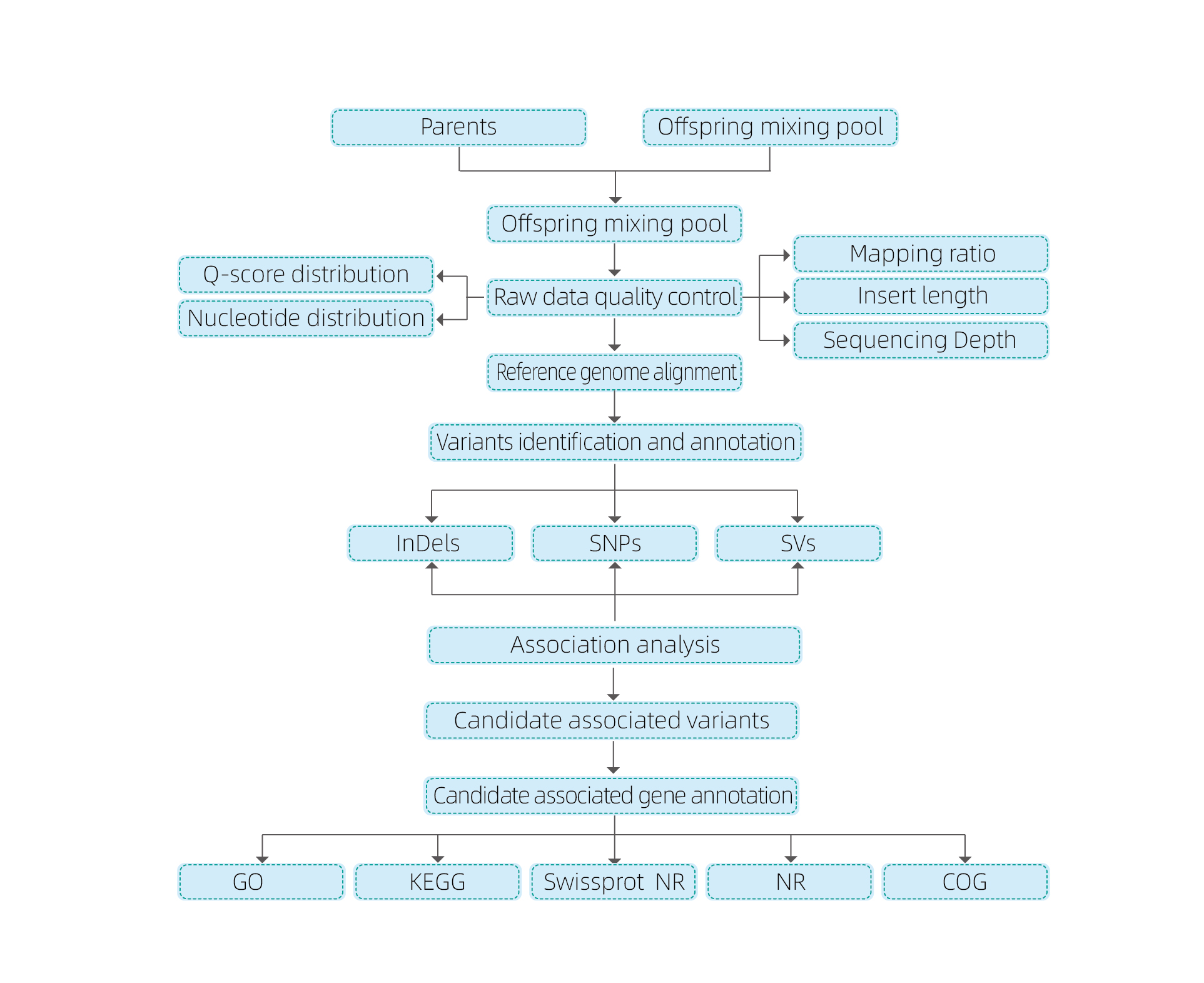
Ama-Bioinformatics ahlaziya
● Ukuvuselela okuphelele kofuzo
● Ukucutshungulwa kwedatha
● I-SNP / Indel Calling
● Ukuhlolwa kwesifunda esifundeni
● Isichasiselo se-gene yofuzo
Izidingo zesampula nokulethwa
Izidingo zesampula:
I-nucleotides:
| isampula le-gdna | Isampula yezicubu |
| Ukuhlushwa: ≥30 ng / μl | Izitshalo: 1-2 g |
| Inani: ≥2 μg (Volummun ≥15 μl) | Izilwane: 0.5-1 g |
| Ukumsulwa: OD260 / 280 = 1.6-2.5 | Igazi eliphelele: 1.5 ml |
Ukugeleza komsebenzi wesevisi

Idizayini yokuhlola

Ukulethwa kwesampula

Ukukhishwa kwe-RNA

Ukwakhiwa Kwelabhulali

Ukulandelana

Ukuhlaziywa kwedatha

Services After-Sale
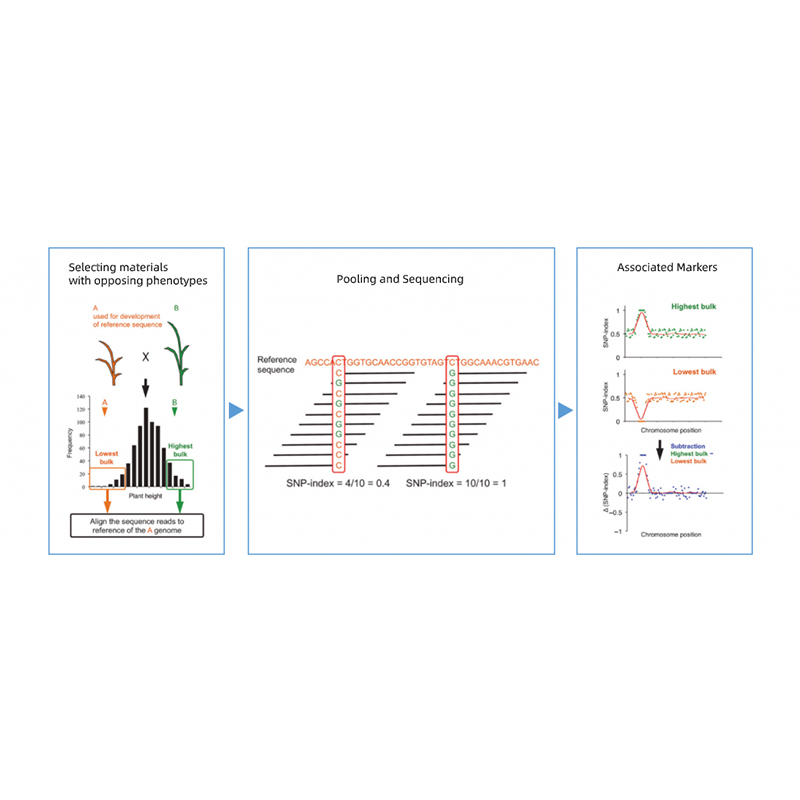
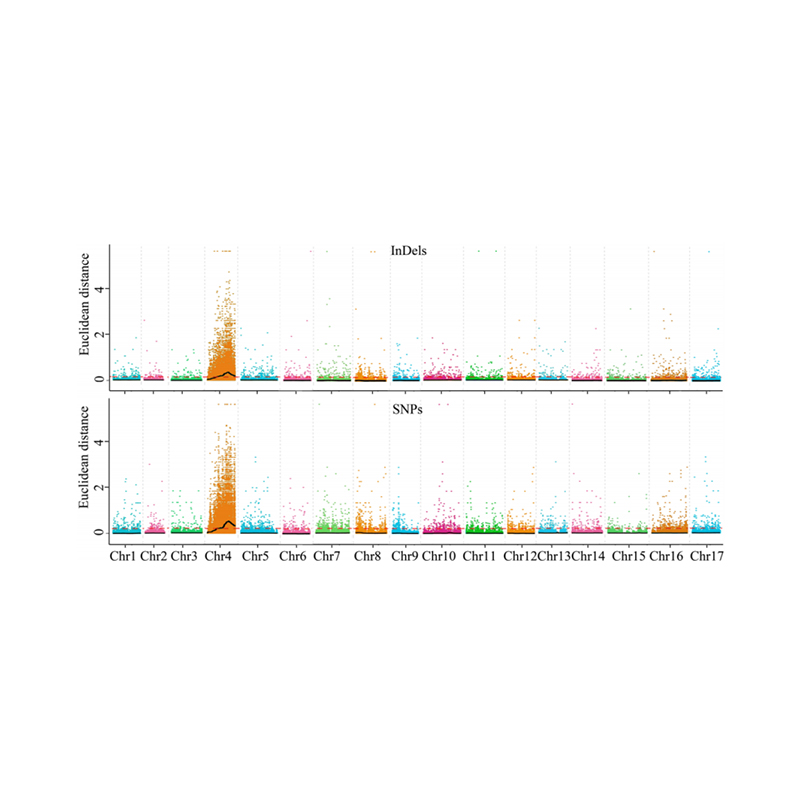
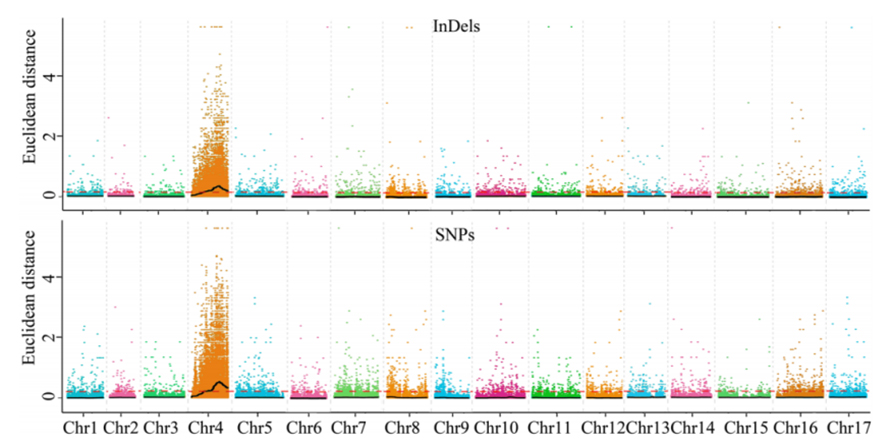
Isisekelo sokuhlaziywa kwe-1.Association ebangeni lase-Euclidean Kulesi sibalo esilandelayo
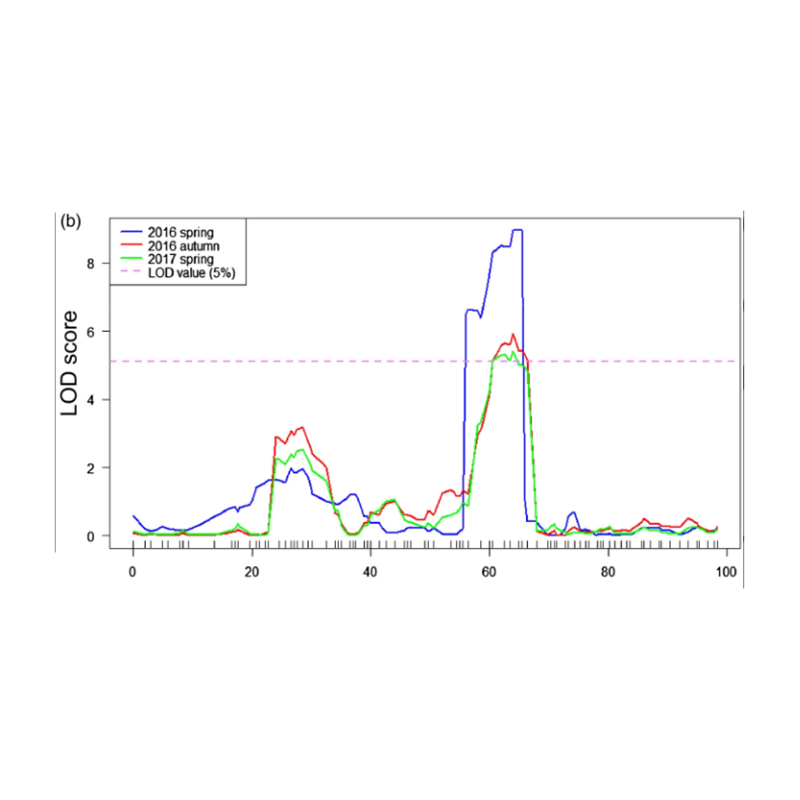
I-X-axis: Inombolo ye-Chromosome; I-dot ngayinye imele inani le-snp. Umugqa omnyama uhambelana nenani elifanelekile. Inani eliphakeme eliphakeme libonisa ubudlelwane obukhulu phakathi kwesiza kanye ne-phenotype. Umugqa we-Red Dash umele umkhawulo wenhlangano ebalulekile.
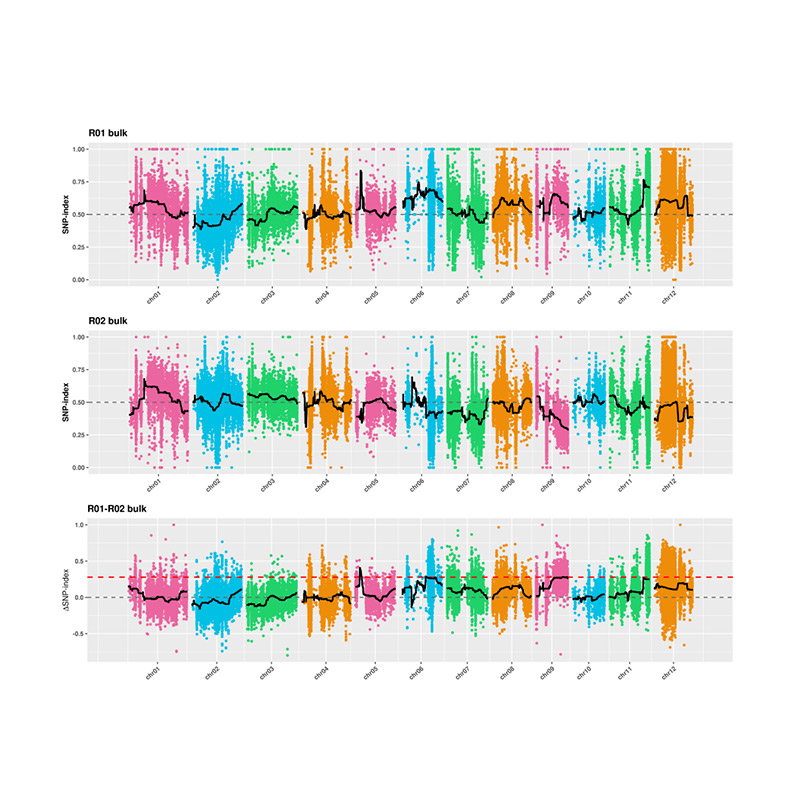
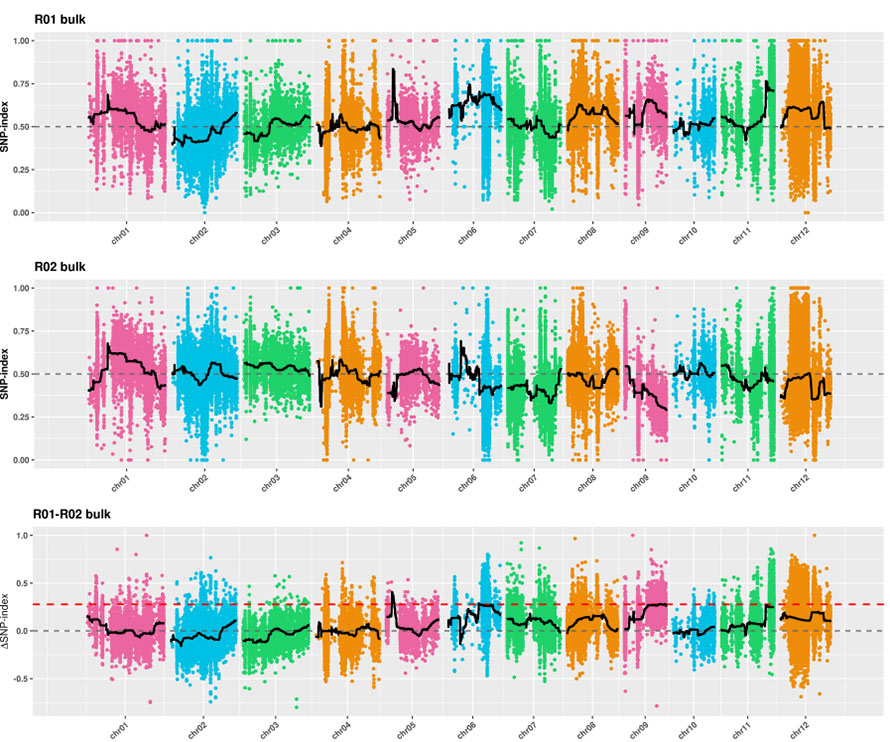
Ukuhlaziywa kwe-2.Association kusekelwe akukho nkomba ye-SNP-Index
I-X-axis: Inombolo ye-Chromosome; I-DOT ngayinye imele inani lenkomba ye-SNP. Umugqa omnyama ume ngenani le-SNP-Index. Okukhulu inani, kubaluleke kakhulu inhlangano.
Icala leBMK
I-Locus ye-Local-Efficial Fetital Fnl7.1 I-Flote WNSROGEGENEIS IPROTEIN enkulu ehlotshaniswa nobude bentamo yezithelo ekhukhamba
Ishicilelwe: I-FRIT BIOTECHnology Journal, 2020
Isu lokulandelana:
Bazali (Jin5-508, yn): Yonke indawo yokuphumula yofuzo i-34 × no 20 ×.
Amachibi e-DNA (ama-50 akhulelwe ama-50 futhi angama-50 amafushane): Ukubuyiselwa kabusha kwe-61 × no-52 ×
Imiphumela Esemqoka
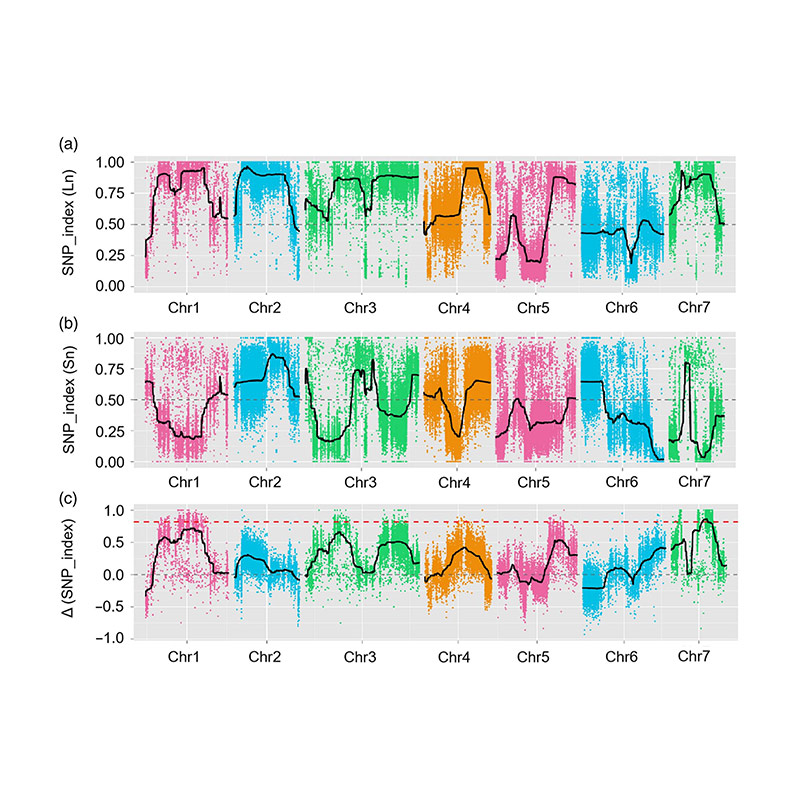
Kulolu cwaningo, ukwahlukanisa inani labantu (F2 ne-F2: 3) kwenziwa ngokuwela umugqa wekhukhamba omude JIN5-508 kanye nentamo ye-yn emfushane. Amachibi amabili e-DNA akhiwa ngabantu abangu-50 abade bentamo ende nabangu-50 abantu abanentamo abafushane. I-QTL enkulu ye-QTL ikhonjwe ku-CHR07 ngokuhlaziywa kwe-BSA kanye nemephu yendabuko yendabuko. Isifunda sokhetho saphinde sancishiswa phansi ngemephu enhle, ukucaciswa kwezichasiso zofuzo kanye nokuhlolwa kwe-transgenic, okuveze uhlobo olusemqoka ekulawuleni ubude bentambo, i-CSFNL7.1. Ngaphezu kwalokho, i-polymorphism ku-CSFNL7.1 Region Region Region yatholakala ihlobene nenkulumo ehambisanayo. Ukuhlaziywa okwengeziwe kwe-phylogenetic kuphakanyiswe ukuthi i-FNL7.1 I-Locus kungenzeka ukuthi yasungulwa e-India.
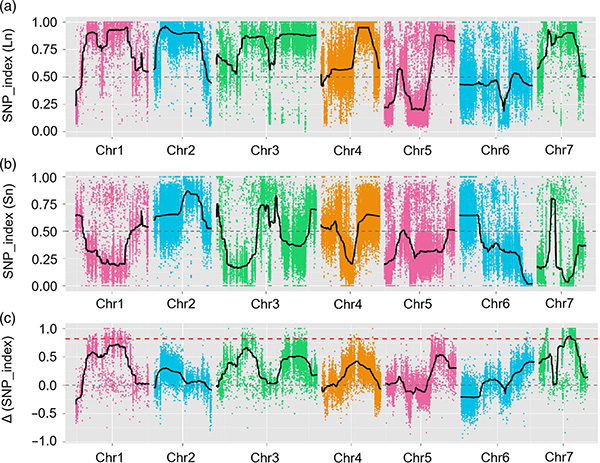 Imephu ye-QTL ekuhlaziyweni kwe-BSA ukukhomba isifunda sokhetho oluhlotshaniswa nobude bemomo yekhukhamba | 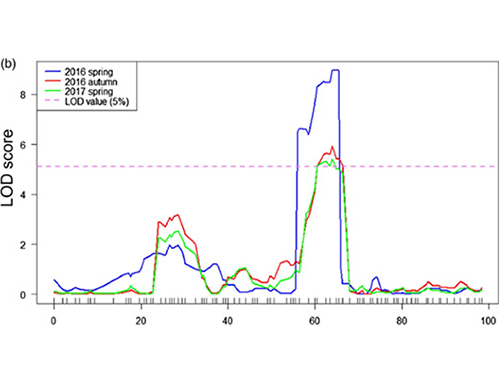 Amaphrofayili weLod amakhukhamba ubude qtl akhonjwe ku-CH07 |
UXu, X., et al. "I-Locus ye-Locutital ye-Majour-Found Locus FNL7.1.1 Ifaka amaprotheni amaningi we-mburry anlogenes ahlotshaniswa nobude bentamo yezithelo ekhukhamba." Plant biotechnology Journal 18.7 (2020).