Specific-Locus Amplified Fragment Sequencing (SLAF-Seq)
Zvimiro Zvebasa
● Sequencing paNovaSeq nePE150.
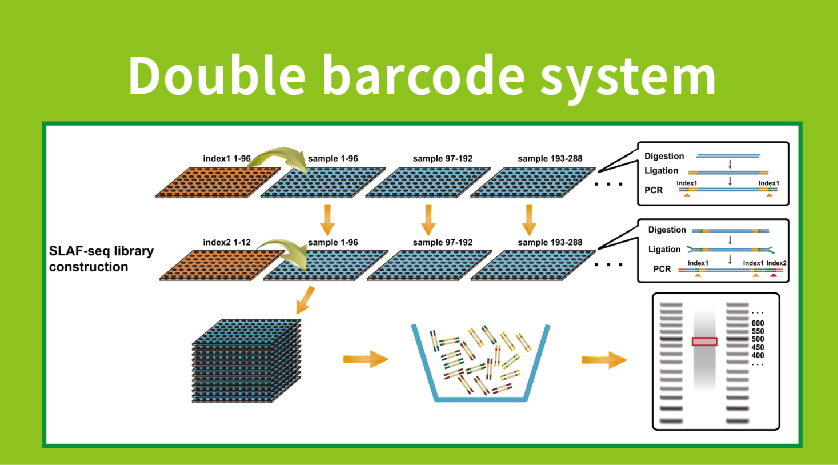
● Kugadzirira raibhurari ine mabhakodhi maviri, zvichiita kuti pave nekubatana kwemasamples anopfuura 1000.
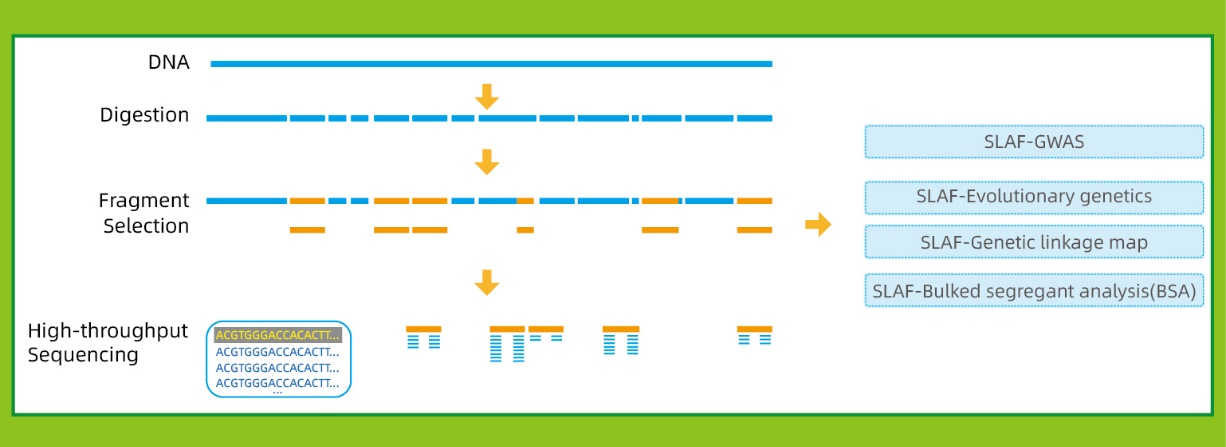
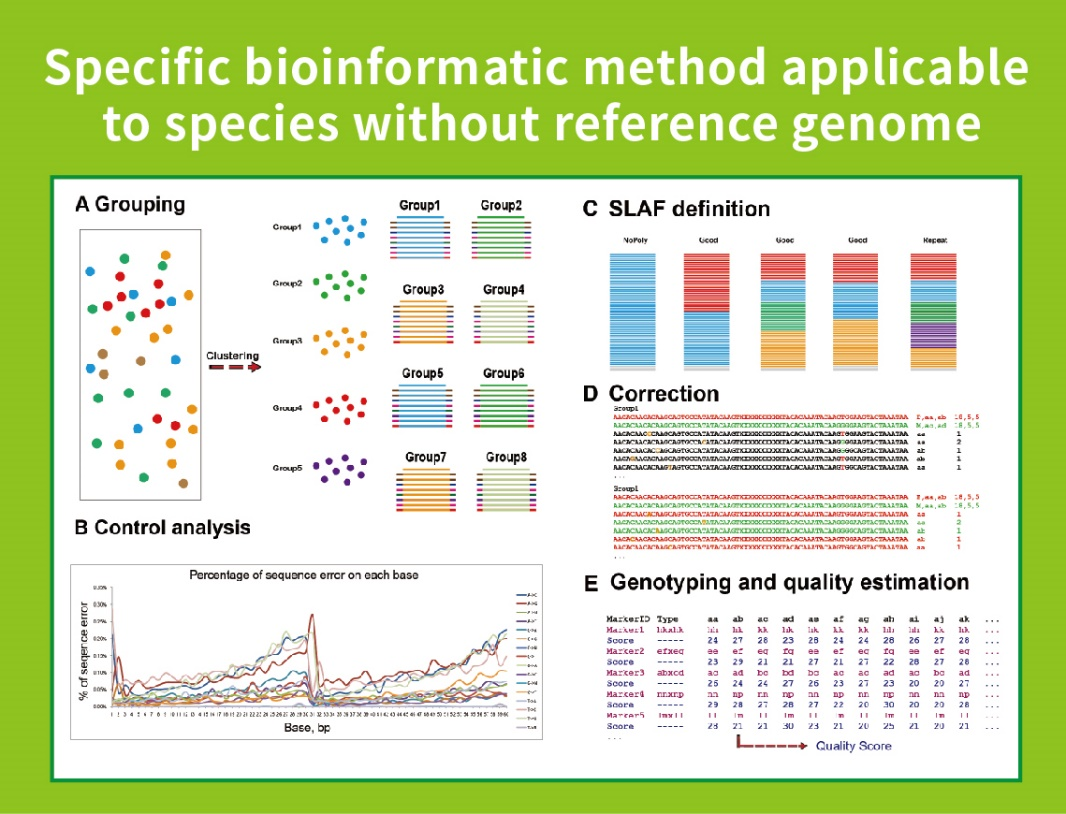
● Nzira iyi inogona kushandiswa kana kuti isina referensi genome, nemapaipi akasiyana ebioinformatic pachiitiko chimwe nechimwe:
Nereferenzi genome: SNP uye InDel kuwanikwa
Pasina referensi genome: sample clustering uye SNP kuwanikwa
● Mumu silicopre-dhizaini nhanho yakawanda inorambidza enzyme musanganiswa inoongororwa kuti iwane ayo anogadzira yunifomu yekugovera yeSLAF tag pamwe nejenome.
● Munguva ye-pre-experiment, masanganiswa matatu ee enzyme anoedzwa mumasampuli matatu kuti abudise maraibhurari 9 eSLAF, uye ruzivo urwu runoshandiswa kusarudza musanganiswa wakakwana wekurambidza enzyme yepurojekiti.
Basa Rinobatsira
●High Genetic Marker Discovery: Kubatanidza yepamusoro-soro kaviri barcode system inobvumira kutevedzana panguva imwe chete yehuwandu hwevanhu, uye locus-specific amplification inowedzera kushanda, kuve nechokwadi kuti nhamba dzemategi dzinosangana nezvinodiwa zvakasiyana-siyana zvemibvunzo yakasiyana-siyana yekutsvakurudza.
● Kutsamira kwakaderera paGenome: Inogona kuiswa kune zvipenyu zvine kana isina referensi genome.
●Flexible Scheme Dhizaini: Single-enzyme, dual-enzyme, multi-enzyme digestion, uye marudzi akasiyana-siyana ema enzymes anogona kusarudzwa kuti aenderane nezvinangwa zvakasiyana zvekutsvagisa kana marudzi. Themu silicopre-dhizaini inoitwa kuti ive nechokwadi cheiyo enzyme dhizaini.
● Kubudirira Kwepamusoro muEnzymatic Digestion: Kuitisa kwemu silicopre-design uye pre-experiment yakavimbiswa dhizaini yakakwana pamwe nekugoverwa kwema tag eSLAF pachromosome (1 SLAF tag/4Kb) uye yakaderedzwa kudzokorora kutevedzana (<5%).
●Hunyanzvi Hwakawanda: Chikwata chedu chinounza hupfumi hweruzivo kupurojekiti yega yega, iine rekodhi yekuvhara anopfuura zviuru zvishanu zveSLAF-Seq mapurojekiti pamazana emarudzi, zvinosanganisira zvinomera, zvinoyamwisa, shiri, tumbuyu, uye zvipenyu zvemumvura.
● Kuzvigadzira pachako Bioinformamatic Workflow: BMKGENE yakagadzira yakasanganiswa bioinformatic workflow yeSLAF-Seq kuve nechokwadi chekuvimbika uye huchokwadi hwekupedzisira kuburitsa.
Sevhisi Zvinotsanangurwa
| Rudzi rwekuongorora | Yakakurudzirwa huwandu hwevanhu | Sequencing strategy | |
| Hudzamu hwema tag sequencing | Tag nhamba | ||
| Genetic Maps | 2 vabereki uye> 150 vana | Vabereki: 20x WGS Kuwedzera: 10x | Genome size: <400 Mb: WGS inokurudzirwa <1Gb: 100K tags 1-2Gb :: 200K tags > 2Gb: 300K tags Max 500k tags |
| Genome-Wide Association Studies (GWAS) | ≥200 samples | 10x | |
| Genetic Evolution | ≥30 samples, ine> gumi samples kubva kuboka rega rega | 10x | |
Zvinodiwa Zvebasa
Kuisa pfungwa ≥ 5 ng/µL
Total mari ≥ 80 ng
Nanodrop OD260/280=1.6-2.5
Agarose gel: hapana kana kushomeka kushomeka kana kusvibiswa
Yakakurudzirwa Sample Delivery
mudziyo: 2 ml centrifuge chubhu
(Kune mazhinji emasampuli, isu tinokurudzira kuti usachengetedze mu ethanol)
Sample labeling: Samples dzinofanirwa kunyorwa zvakajeka uye dzakafanana neyakatumirwa ruzivo ruzivo fomu.
Shipment: Dry-ice: Samples dzinofanirwa kurongedzerwa mumabhegi kutanga uye kuvigwa mune yakaoma-aizi.
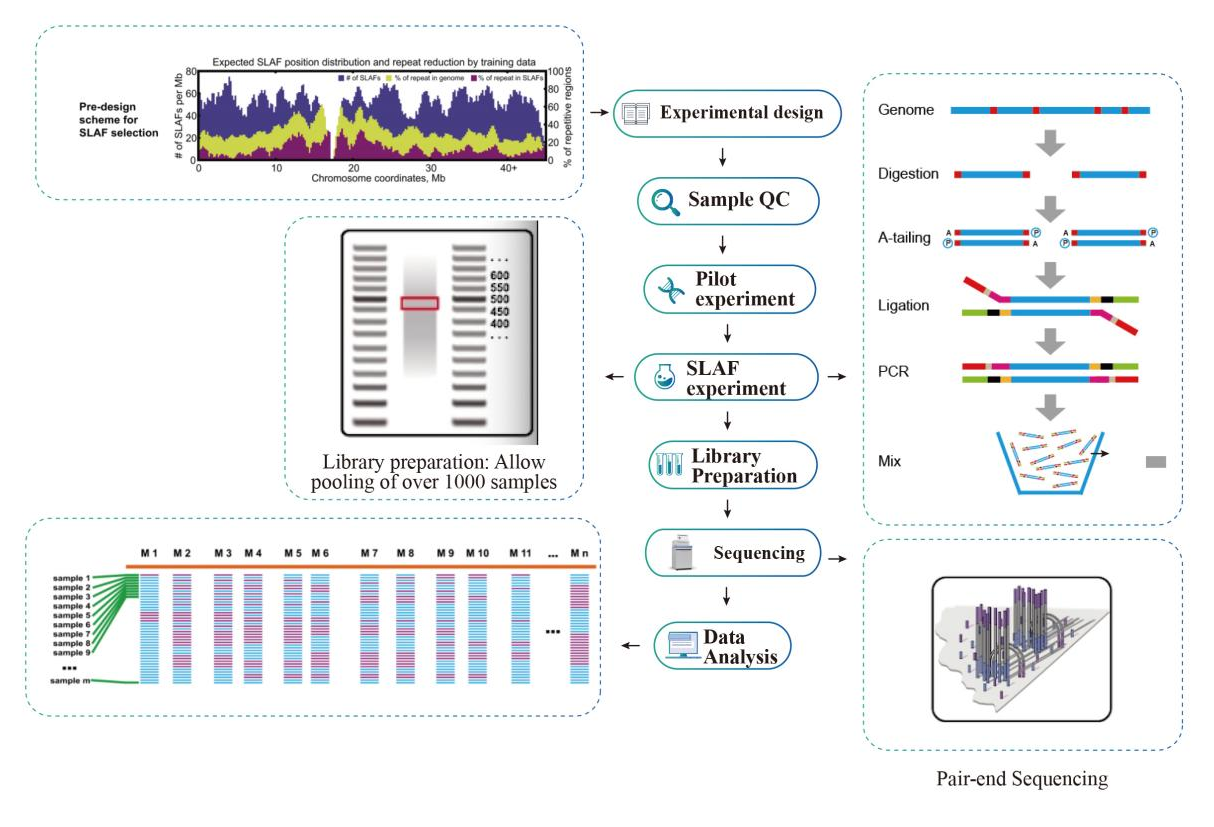
Service Workflow






Muenzaniso QC
Pilot experiment
SLAF-kuedza
Kugadzirira Raibhurari
Sequencing
Data Analysis
Mushure mekutengesa Services
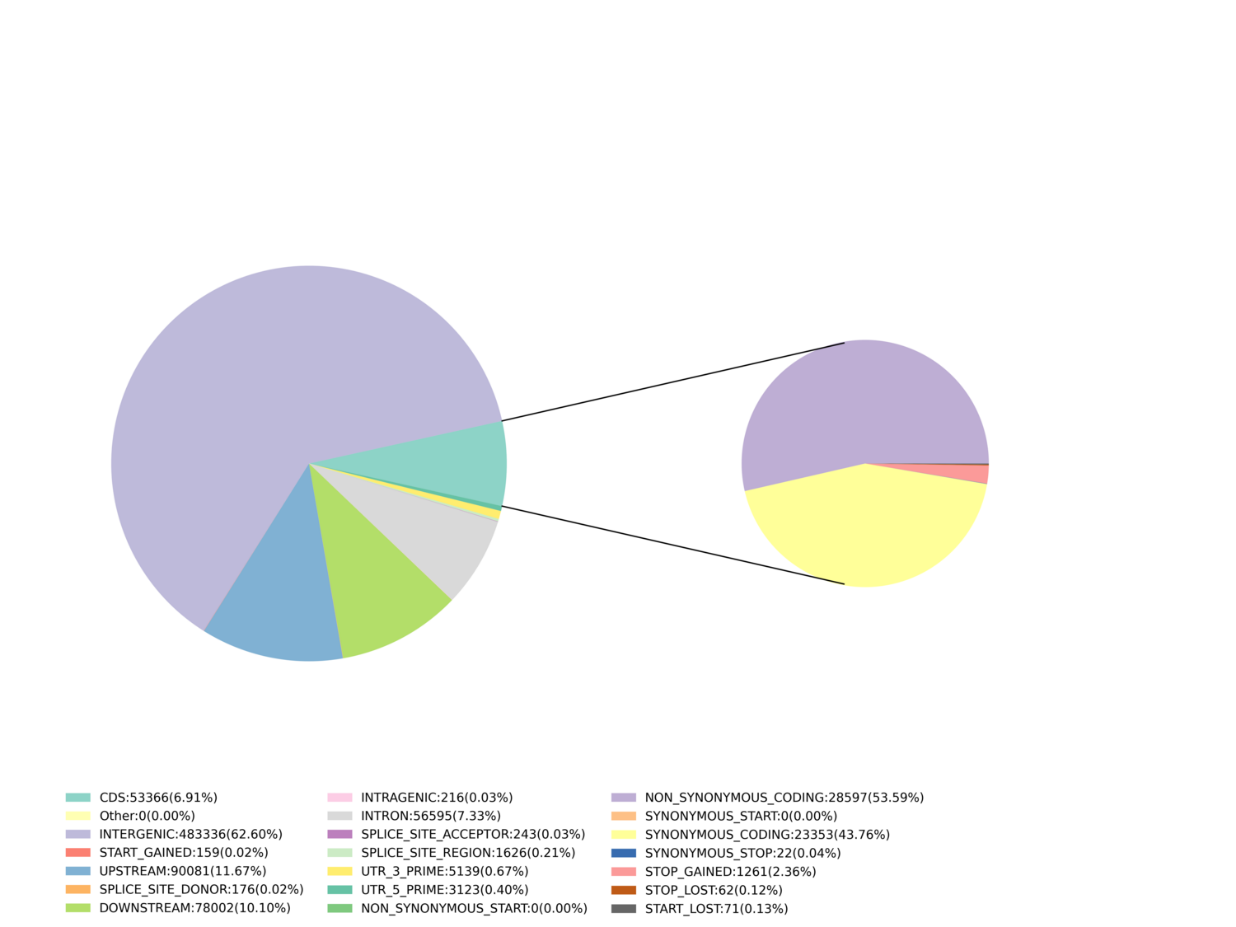
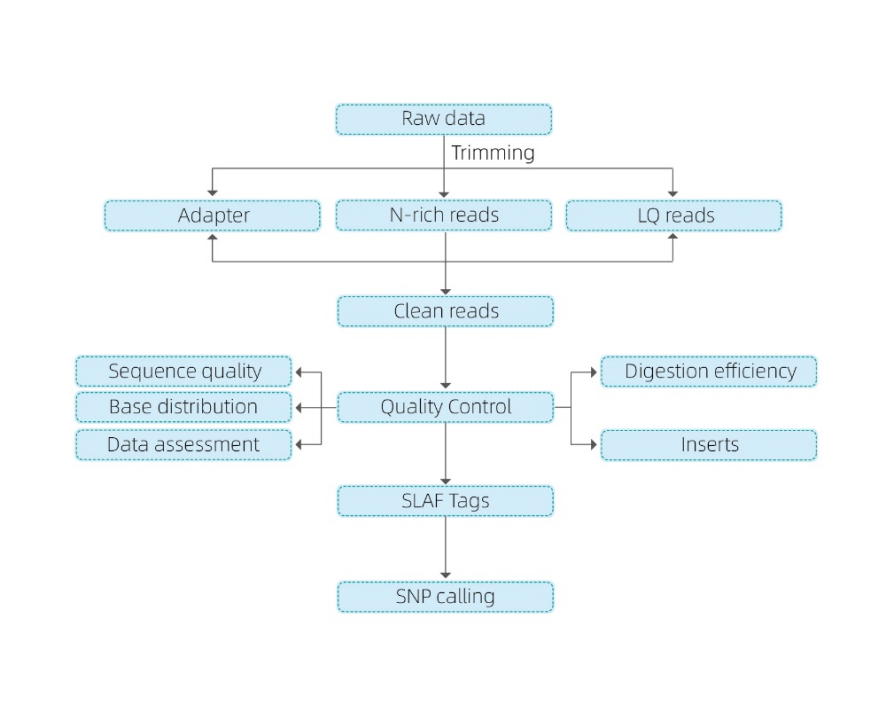 Inosanganisira ongororo inotevera:
Inosanganisira ongororo inotevera:
- Sequencing data QC
- SLAF tag kuvandudzwa
Mepu yekutarisa genome
Pasina referensi genome: clustering
- Ongororo yeSLAF tags.: manhamba, kugovera pane genome
- Mucherechedzo kuwanikwa: SNP, InDel, SNV, CV kufona uye chirevo
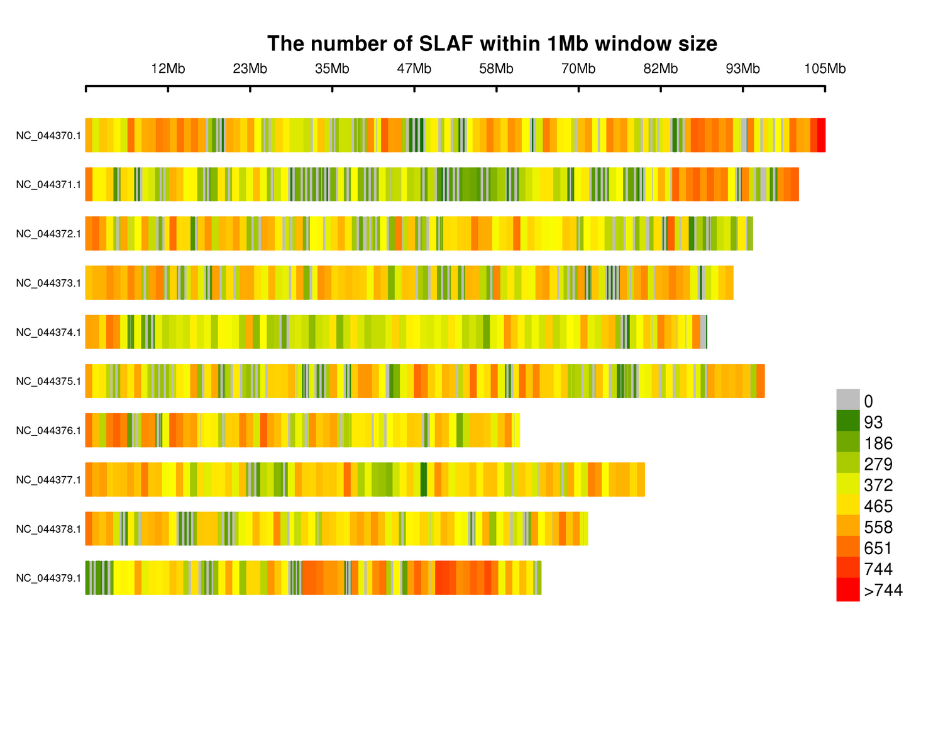
Kugoverwa kwematagi eSLAF pamakromosomes:
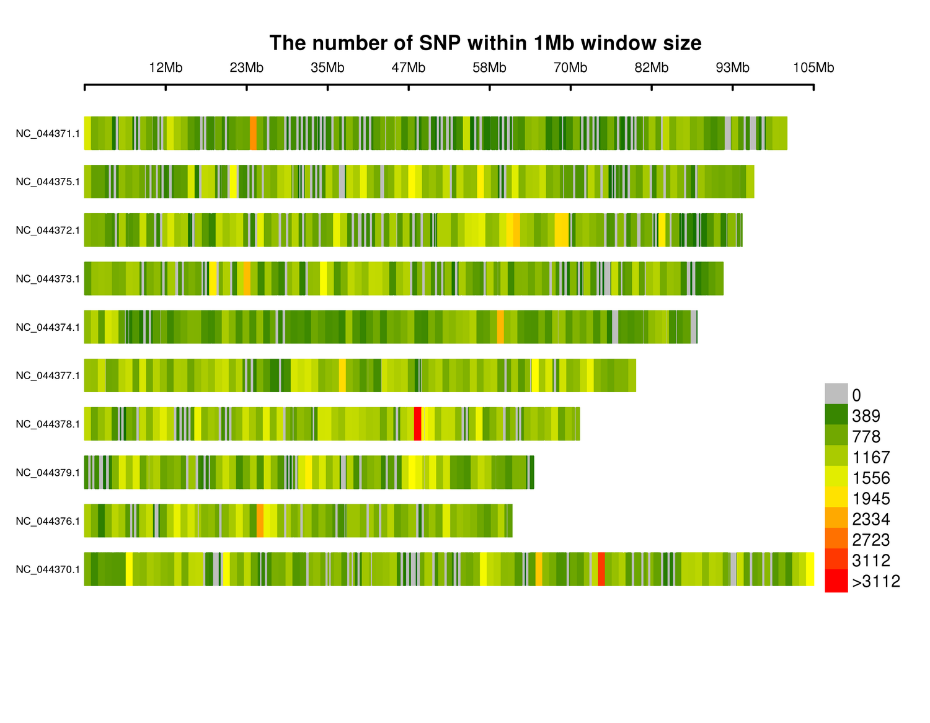
Kugoverwa kweSNPs pamakromosomes:
| Gore | Journal | IF | Title | Applications |
| 2022 | Kutaurirana kwakasikwa | 17.694 | Genomic nheyo yegiga-chromosomes uye giga-genome yemuti peony Paeonia ostii | SLAF-GWAS |
| 2015 | New Phytologist | 7.433 | Domestication footprints anchor genomic regions of agronomic value in soya beans | SLAF-GWAS |
| 2022 | Chinyorwa che Advanced Research | 12.822 | Genome-wide artificial introgressions yeGossypium barbadense muG. hirsutum ratidza yepamusoro loci yekuvandudza panguva imwe chete yekotoni fiber mhando uye goho maitiro | SLAF-Evolutionary genetics |
| 2019 | Molecular Plant | 10.81 | Population Genomic Analysis uye De Novo Assembly Inozivisa Mavambo eWeedy Mupunga semutambo weEvolutionary | SLAF-Evolutionary genetics |
| 2019 | Nature Genetics | 31.616 | Genome sequence uye genetic diversity of the common carp, Cyprinus carpio | Mepu yeSLAF-Linkage |
| 2014 | Nature Genetics | 25.455 | Iyo genome yenzungu yakarimwa inopa nzwisiso mune legume karyotypes, polyploid. evolution uye kurima zvirimwa. | Mepu yeSLAF-Linkage |
| 2022 | Plant Biotechnology Journal | 9.803 | Kuzivikanwa kwe ST1 kunoratidza sarudzo inosanganisira kurovera kwembeu morphology uye zviri mukati memafuta panguva yekudyara soya bean | SLAF-Mucherechedzo kuvandudza |
| 2022 | International Journal yeMolecular Sayenzi | 6.208 | Kuzivikanwa uye DNA Mucherechedzo Kukudziridzwa kweGorosi-Leymus mollis 2Ns (2D) Disomic Chromosome Kutsiva | SLAF-Mucherechedzo kuvandudza |
| Gore | Journal | IF | Title | Applications |
| 2023 | Miganhu mune sainzi yezvirimwa | 6.735 | QTL mepu uye ongororo yezvinyorwa zveshuga panguva yekuibva kwemichero yePyrus pyrifolia | Genetic Map |
| 2022 | Plant Biotechnology Journal | 8.154 | Kuzivikanwa kwe ST1 kunoratidza sarudzo inosanganisira kurovera kwembeu morphology uye zvemukati mafuta panguva yekupfuya soya.
| SNP kufona |
| 2022 | Miganhu mune sainzi yezvirimwa | 6.623 | Genome-Wide Association Mepu yeHulless Barely Phenotypes mune Drought Environment.
| GWAS |