Genome-wide Association Analysis
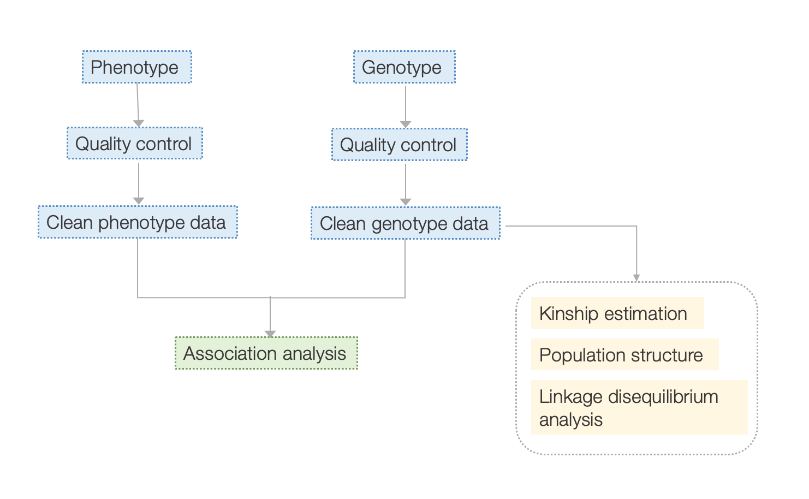
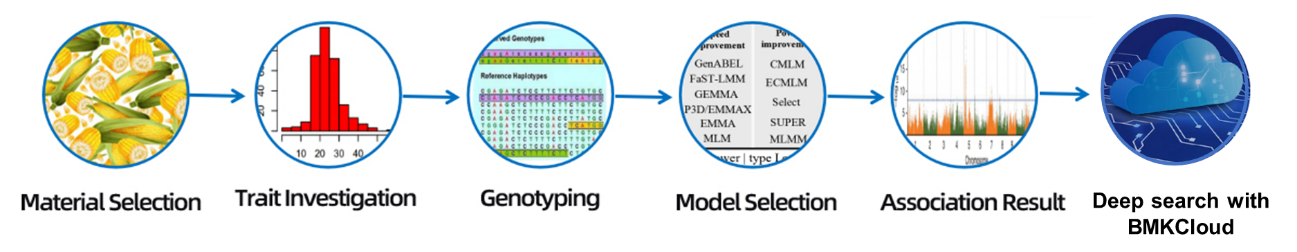
Workflow
Basa Rinobatsira
●Hunyanzvi Hwakawanda uye zvinyorwa zvekushambadzira: iine ruzivo rwakaunganidzirwa muGWAS, BMKGene yapedza mazana emapurojekiti ezvipenyu mukutsvagisa kwehuwandu hweGWAS, yakabatsira vaongorori kuburitsa zvinopfuura zvinyorwa zve100, uye iyo cumulative impact factor yakasvika mazana mashanu.
● Comprehensive bioinformatics analysis: kufambiswa kwebasa kunosanganisira SNP-hunhu hwekusangana ongororo, kuendesa seti yevamiriri majini uye yavo inoenderana inoshanda annotation.
●Yakanyanya hunyanzvi bioinformatics timu uye pfupi yekuongorora kutenderera: ine ruzivo rwakakura mukuongorora kwepamusoro genomics, timu yeBMKGene inopa ongororo yakazara nenguva inokurumidza yekuchinja.
●Post-Sales Support:Kuzvipira kwedu kunowedzera kupfuura kupera kweprojekiti nemwedzi mitatu mushure mekutengesa sevhisi nguva. Munguva ino, tinopa kutevedza kweprojekiti, rubatsiro rwekugadzirisa matambudziko, uye maQ&A masesheni kugadzirisa chero mibvunzo ine chekuita nemhedzisiro.
Sevhisi yakatarwa uye zvinodiwa
| Rudzi rwekutevedzana | Yakakurudzirwa huwandu hwevanhu | Sequencing strategy | Nucleotide zvinodiwa |
| Yese Genome Sequencing | 200 samples | 10x | Kuisa pfungwa: ≥ 1 ng/ µL Total mari≥ 30ng Yakaganhurirwa kana isina kudzikisira kana kusvibiswa |
| Specific-Locus Amplified Fragment (SLAF) | Kudzika kweTag: 10x Nhamba yematagi: <400 Mb: WGS inokurudzirwa <1Gb: 100K tags 1Gb > 2Gb: 300K tags Max 500k tags | Kuisa pfungwa ≥ 5 ng/µL Total mari ≥ 80 ng Nanodrop OD260/280=1.6-2.5 Agarose gel: hapana kana kushomeka kushomeka kana kusvibiswa
|
Kusarudzwa kwezvinhu



Mhando dzakasiyana, subspecies, landraces / genebanks / mhuri dzakasanganiswa / zviwanikwa zvemusango
Mhando dzakasiyana, subspecies, landraces
Half-sib family/full-sib family/wild resources
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

RNA kubviswa

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
Inosanganisira ongororo inotevera:
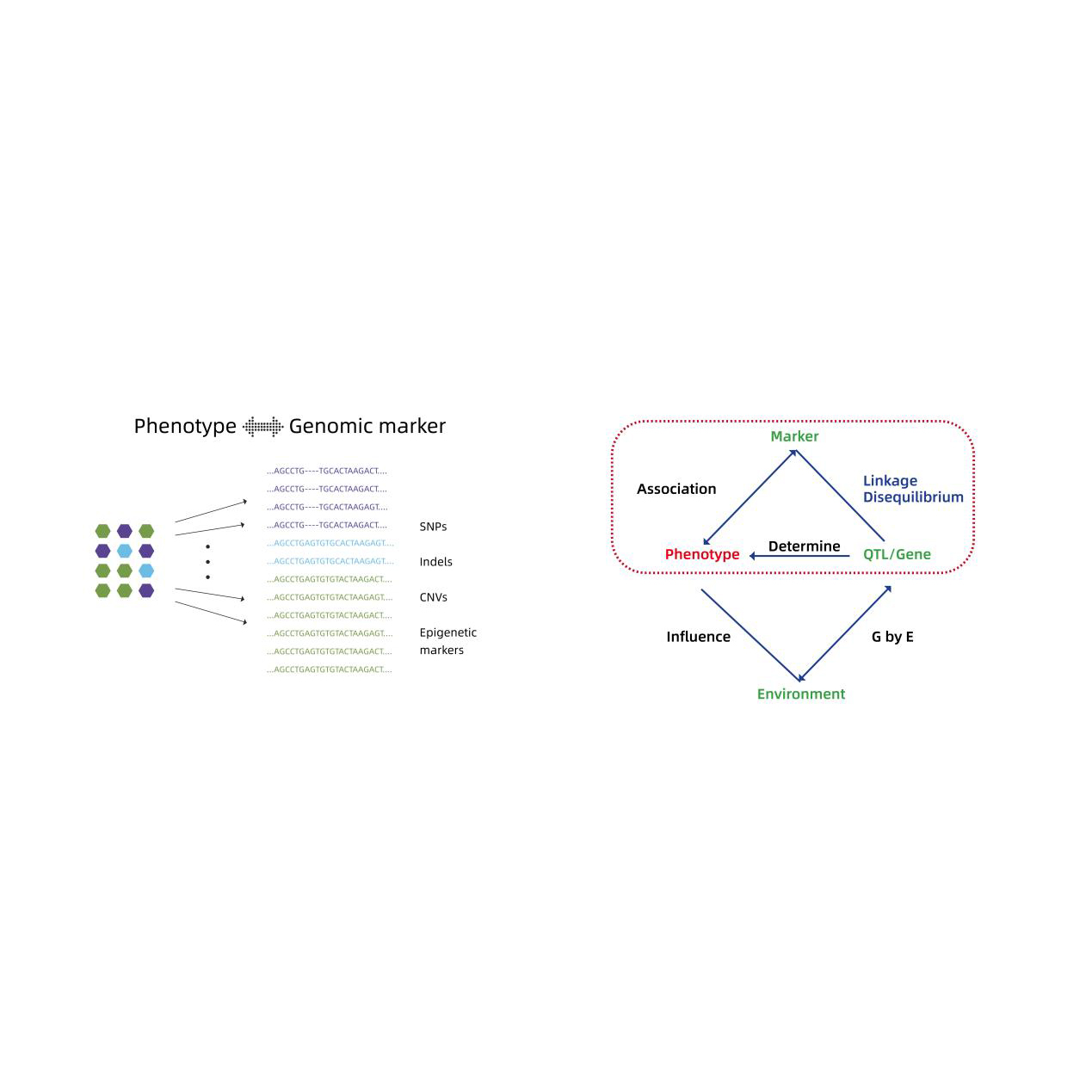
- Genome-wide association analysis: LM, LMM, EMMAX, FASTLMM modhi
- Tsanangudzo inoshanda yemajini emumiriri
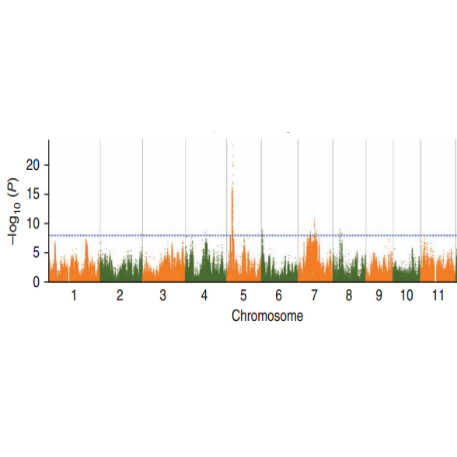
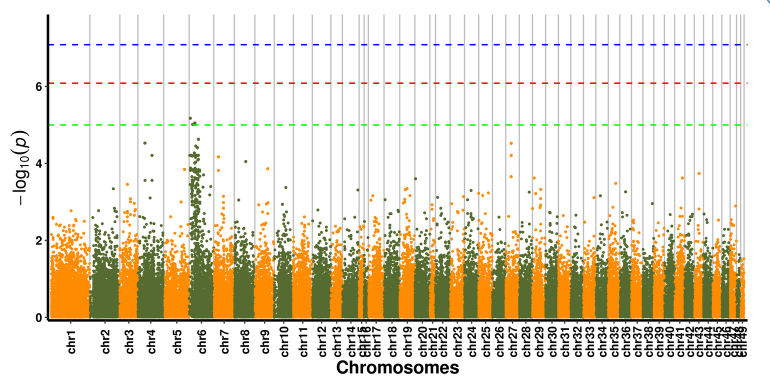
SNP-trait association analysis - Manhattan plot
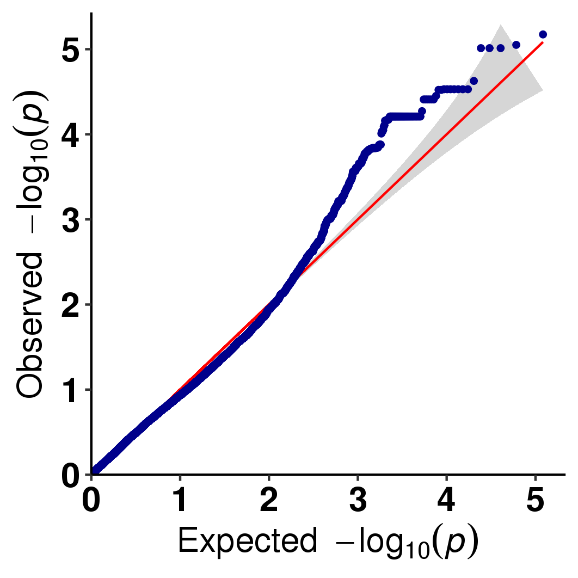
SNP-trait association analysis - QQ plot
Ongorora kufambira mberi kunofambiswa neBMKGene's de GWAS masevhisi kuburikidza neakaunganidzwa muunganidzwa wezvinyorwa:
Lv, L. nevamwe. (2023) 'Kunzwisisa kune genetic hwaro hwekushivirira kweammonia mu razor clam Sinonovacula constricta ne genome-wide association kudzidza',Aquaculture, 569, p. 739351. doi: 10.1016/J.AQUACULTURE.2023.739351.
Li, X nevamwe. (2022) 'Multi-omics analyzes ye398 foxtail millet accessions inoratidza genomic matunhu ane chekuita nemusha, metabolite maitiro, uye anti-kuzvimba mhedzisiro',Molecular Plant, 15(8), mapeji 1367–1383. doi: 10.1016/j.molp.2022.07.003.
Li, J. nevamwe. (2022) 'Genome-Wide Association Mepu yeHulless Barely Phenotypes muKusanaya Kwemvura',Frontiers muPlant Sayenzi, 13, p. 924892. doi: 10.3389/FPLS.2022.924892/BIBTEX.
Zhao, X. nevamwe. (2021) 'GmST1, iyo inoisa sulfotransferase, inopa kuramba kune soyabean mosaic virus strains G2 uye G3',Plant, Cell & Environment, 44(8), mapeji 2777–2792. doi: 10.1111/PCE.14066.