De novo Fungal Genome Assembly
Zvimiro Zvebasa
Nezvitatu zvinogoneka sarudzo dzekusarudza kubva zvichienderana neinodiwa dhigirii rekuzara kwegenome:
● Draft Genome Option: Pfupi-kuverenga kutevedzana neIllumina NovaSeq PE150.
● Fungal Fine Genome Option:
Genome Survey: Illumina NovaSeq PE150.
Genome Assembly: PacBio Revio (HiFi inoverenga) kana Nanopore PromethION 48.
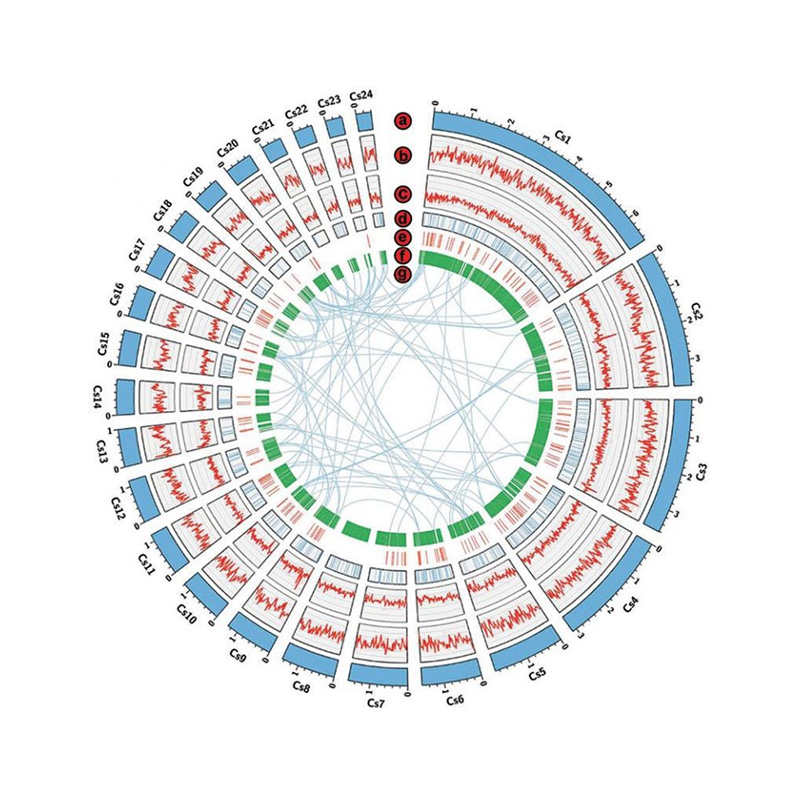
● Chromosome-level Fungal Genome:
Genome Survey: Illumina NovaSeq PE150.
Genome Assembly: PacBio Revio (HiFi inoverenga) kana Nanopore PromethION 48.
Contig anchoring neHi-C assembly.
Basa Rinobatsira
●Multiple Sequencing Strategies Inowanikwa: Nezvinangwa zvakasiyana zvekutsvagisa uye zvinodiwa zvekuzara kwegenome
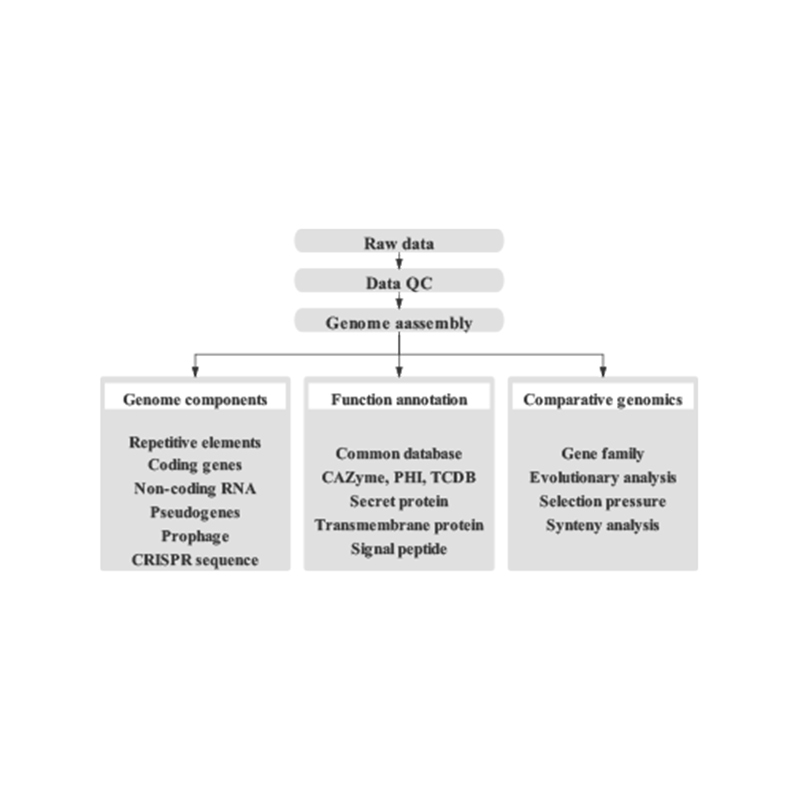
●Zadzisa Bioinformatics Workflow:Izvi zvinosanganisira kusangana kwegenome uye kufanotaura kweakawanda genomic element, inoshanda gene annotation, uye contig anchoring.
●Hunyanzvi Hwakawanda: Nezvinopfuura zviuru gumi nezviviri zvemicrobial genomes zvakaunganidzwa, tinounza anopfuura makore gumi echiitiko, timu yekuongorora ine hunyanzvi, yakazara zvemukati, uye yakanakisa post-kutengesa rutsigiro.
●Post-Sales Support:Kuzvipira kwedu kunowedzera kupfuura kupera kweprojekiti nemwedzi mitatu mushure mekutengesa sevhisi nguva. Munguva ino, tinopa kutevedza kweprojekiti, rubatsiro rwekugadzirisa matambudziko, uye maQ&A masesheni kugadzirisa chero mibvunzo ine chekuita nemhedzisiro.
Sevhisi Zvinotsanangurwa
| Service | Sequencing Strategy | Quality Control |
| Draft Genome | Illumina PE150 100x | Q30≥85% |
| Fine Genome | Genome ongororo: Illumina PE150 50 x Gungano: PacBio HiFi 30x kana Nanopore 100x | contig N50 ≥1Mb (Pacbio Unicellular) contig N50 ≥2Mb (ONT Unicellular) contig N50 ≥500kb (Zvimwe) |
| Chromosome-level genome | Genome ongororo: Illumina PE150 50 x Gungano: PacBio HiFi 30x kana Nanopore 100x Hi-C Gungano 100x | Contig anchoring ratio>90%
|
Zvinodiwa Zvebasa
| Concentration (ng/µL) | Huwandu hwese (µg) | Vhoriyamu (µL) | OD260/280 | OD260/230 | |
| PacBio | ≥20 | ≥2 | ≥20 | 1.7-2.2 | ≥1.6 |
| Nanopore | ≥40 | ≥2 | ≥20 | 1.7-2.2 | 1.0-3.0 |
| Illumina | ≥1 | ≥0.06 | ≥20 | - | - |
Unicellular Fungus: ≥3.5x1010 masero
Macro Fungus: ≥10 g
Kuyerera Kwebasa Rebasa

Muenzaniso wekutumira

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
Inosanganisira Kuongorora Kunotevera:
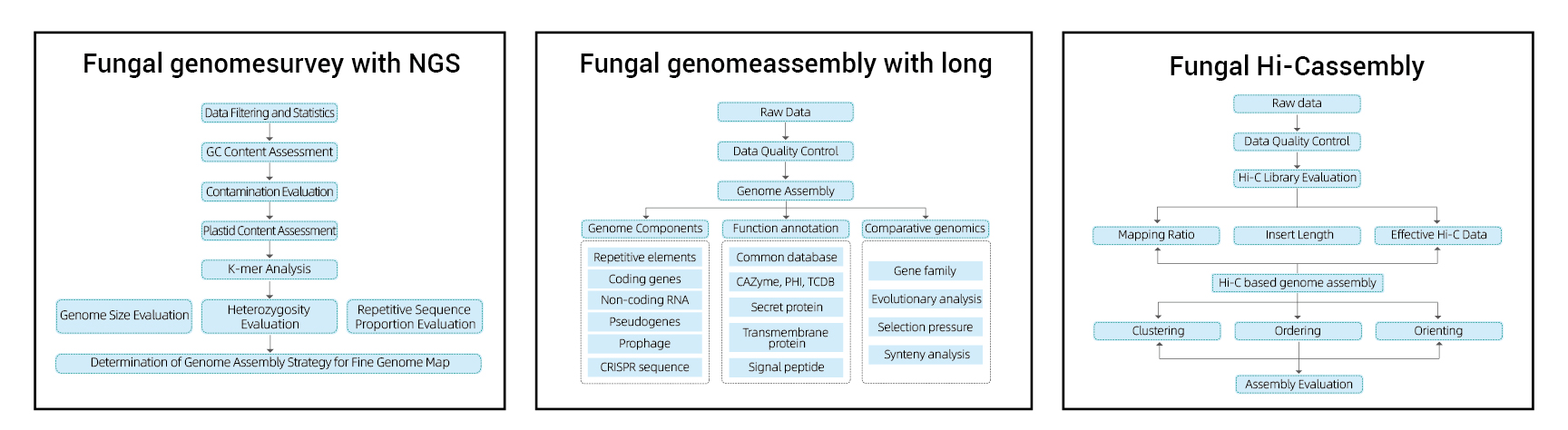
Genome Survey:
- Sequencing data quality control
- Genome estimation: saizi, heterozygosity, kudzokorora zvinhu
Yakanaka Genome Assembly:
- Sequencing Data Quality Control
- De novoAssembly
- Genome Component Analysis: Kufanotaura kweCDS uye akawanda genomic elements
- Inoshanda annotation ine akawanda general dhatabhesi (GO, KEGG, nezvimwewo) uye epamberi dhatabhesi (CARD, VFDB, nezvimwewo)
Hi-C Assembly:
- Hi-C Library Ongororo.
- Contigs anchoring clustering, kuronga uye kutungamira
- Kuongororwa kweHI-C Gungano: Kubva pane referensi genome uye kupisa mepu
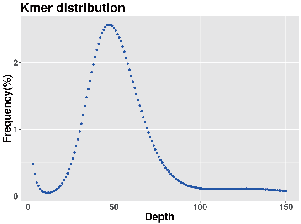
Genome ongororo: k-mer kugovera
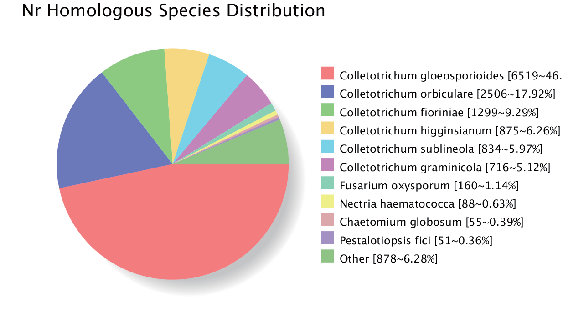
Genome musangano: gene homologous annotation (NR dhatabhesi)
Genome musangano: inoshanda gene annotation (GO)
Ongorora kufambira mberi kunofambiswa neBMKGene's fungal genome musangano masevhisi kuburikidza neakaunganidzwa muunganidzwa wezvinyorwa.
Hao, J. nevamwe. (2023) 'Yakabatanidzwa omic profiling yemishonga mushroom Inotus obliquus pasi pemamiriro akanyura',BMC Genomics, 24(1), map. 1–12. doi: 10.1186/S12864-023-09656-Z/FIGURES/3.
Lu, L. nevamwe. (2023) 'Genome sequencing inoratidza shanduko uye pathogenic nzira dzegorosi rakapinza maziso pathogen Rhizoctonia cerealis',The Crop Journal, 11(2), mapeji 405–416. doi: 10.1016/J.CJ.2022.07.024.
Zhang, H. et al. (2023) 'Genome Zviwanikwa zveMana Clarireedia Zvisikwa Zvinokonzera Dollar Spot pane Diverse Turfgrasses',Plant Disease, 107(3), mapeji 929–934. doi: 10.1094/PDIS-08-22-1921-A
Zhang, SS et al. (2023) 'Genetic uye Molecular Evidence yeTetrapolar Mating System muEdible Mushroom Grifola frondosa',Nyaya yeFungi, 9(10), p. 959. doi: 10.3390/JOF9100959/S1.