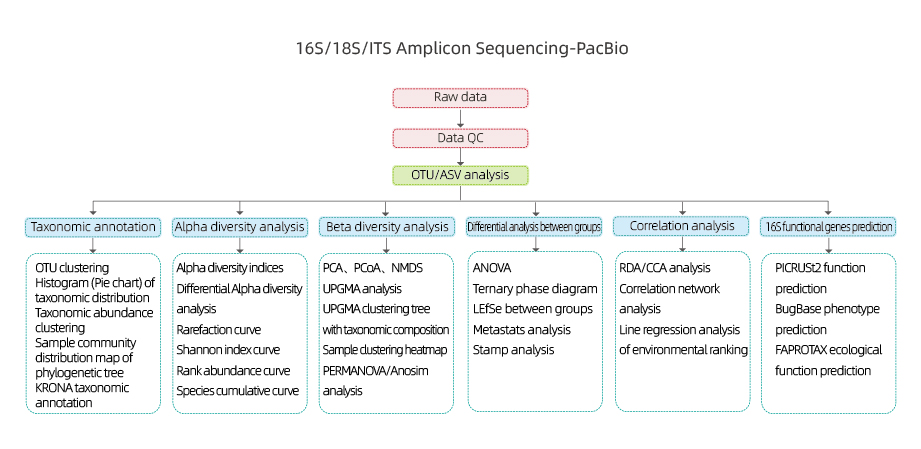
16S/18S/ITS Amplicon Sequencing-PacBio
Zvimiro Zvebasa
● Sequencing platform: PacBio Revio
● Sequencing mode: CCS (HiFi inoverenga)
● Kukwidziridzwa kwenzvimbo inotarirwa inoteverwa nekubatanidza tandem yemaamplicon pamberi peHiFi SMRT bhero raibhurari yekugadzirira.
Basa Rinobatsira
●Yakakwira taxonomic resolution: Than short-amplicon sequencing,kugonesa yakakwirira yeOTU kurongedza mazinga padanho rezvipenyu.
●Yakanyanya kufona base kufona: PacBio CCS mode sequencing (HiFi inoverenga).
●Kuzviparadzanisa nevamwe: Kukurumidza kuzivikanwa kweiyo microbial kuumbwa mune zvakatipoteredza samples.
●Zvakawanda zvinoshanda: Zvidzidzo zvakasiyana siyana zvenharaunda.
●Comprehensive bioinformatic analysis: Iyo yazvino QIIME2 package (yehuwandu nzwisiso mumicrobial ecology) ine ongororo dzakasiyana maererano nedatabase, zvirevo, OTU/ASV.
●Hunyanzvi Hwakawanda: Nezviuru zveamplicon sequencing mapurojekiti anoitwa gore negore, BMKGENE inounza anopfuura makore gumi echiitiko, timu yekuongorora ine hunyanzvi, yakazara zvemukati, uye yakanakisa post-kutengesa rutsigiro.
Sevhisi Zvinotsanangurwa
| Library | Sequencing Strategy | Data inokurudzirwa |
| Amplicon | PacBio Revio | 10/30/50 K tags (CCS) |
Muenzaniso Zvinodiwa
| Concentration (ng/µL) | Huwandu hwese (µg) | Vhoriyamu (µL) |
| ≥5 | ≥0.3 | ≥20 |
Yakakurudzirwa Sample Delivery
Svitsa masampula munitrogen yemvura kwemaawa 3-4 uye chengetedza mune yemvura nitrogen kana -80 dhigirii kusvika kurebesa kwenguva refu. Sample shipping with dry-ice inodiwa.
Kuyerera Kwebasa Rebasa

Muenzaniso wekutumira

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
Inosanganisira ongororo inotevera:
●Raw data quality control
● OTU clustering/De-noise(ASV)
● OTU chirevo
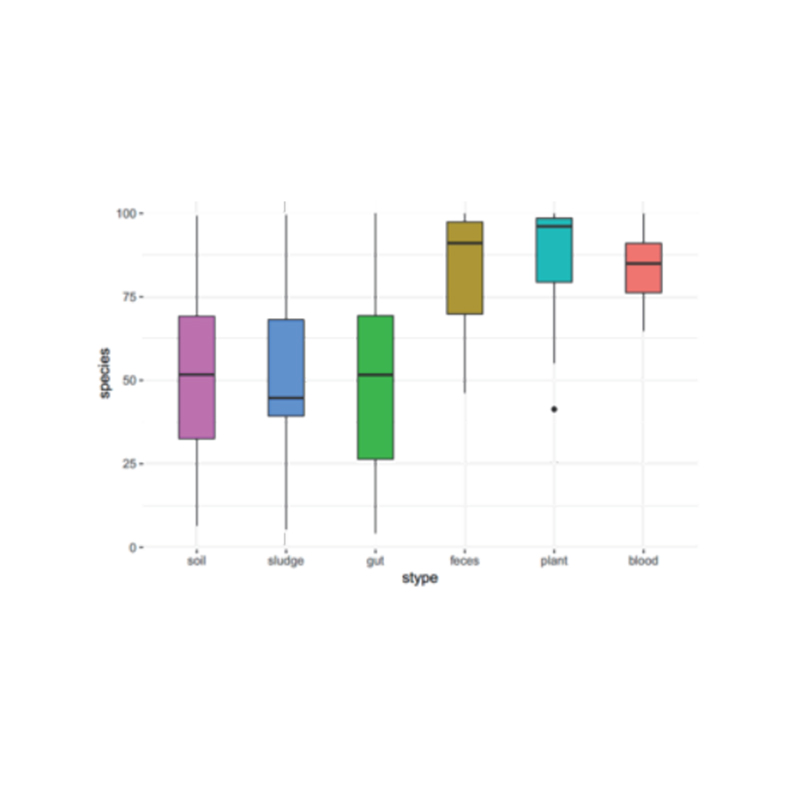
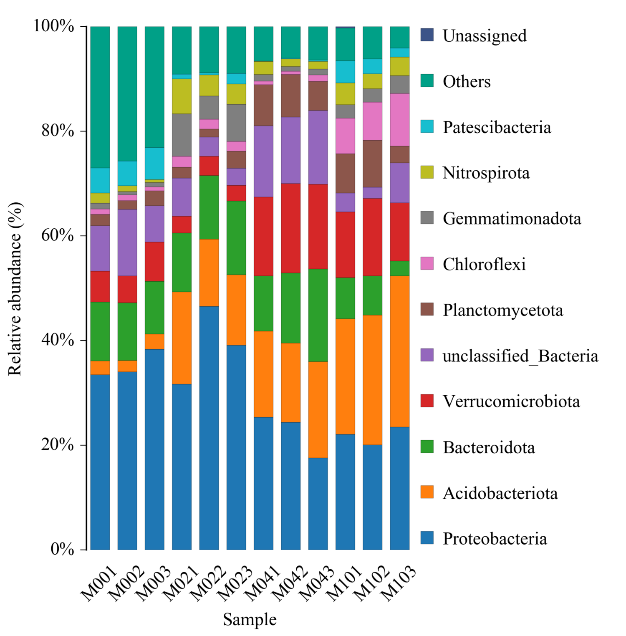
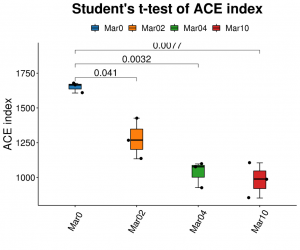
● Alpha diversity analysis: akawanda indexes, kusanganisira Shannon, Simpson uye ACE.
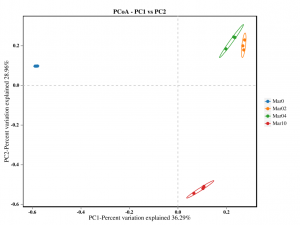
●Kuongorora kweBeta kusiyana-siyana
●Kuongorora pakati peboka
● Correlation analysis: pakati pezvakatipoteredza zvinhu uye OUT kuumbwa uye kusiyana-siyana
● 16S inoshanda gene kufanotaura
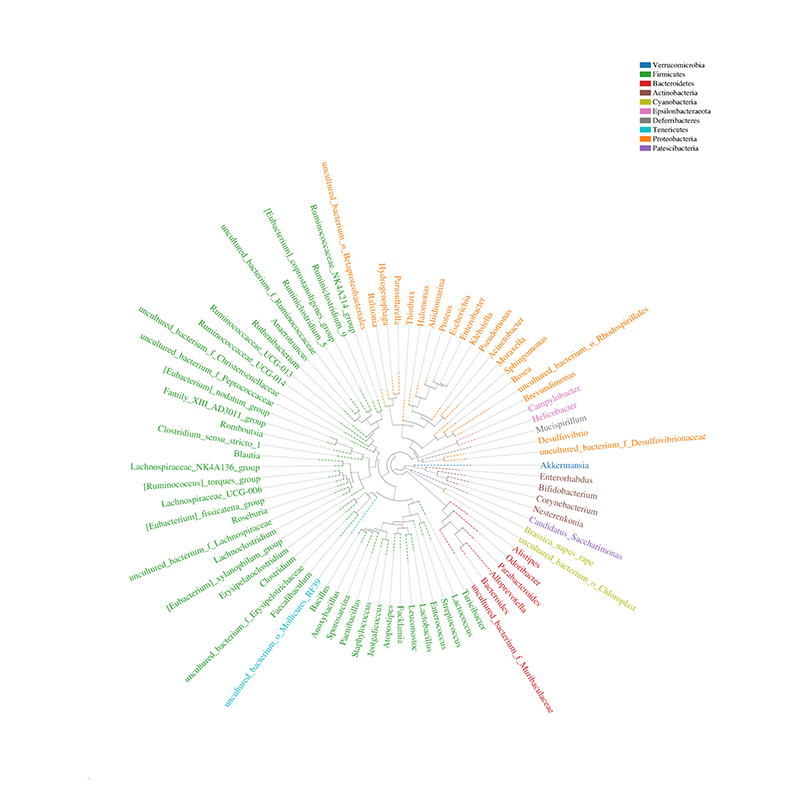
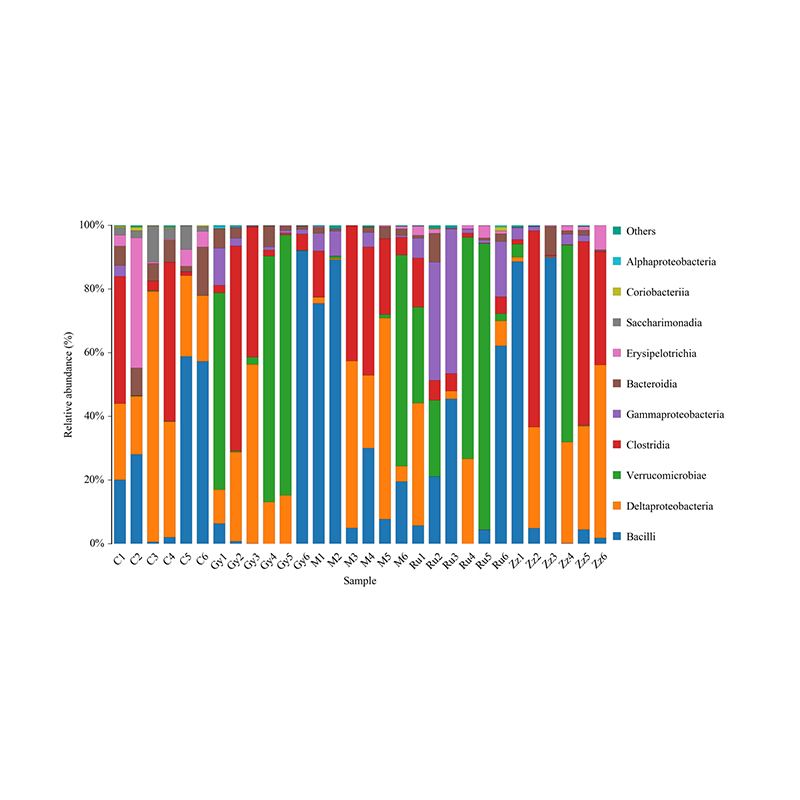
Histogram yekugovera taxonomic
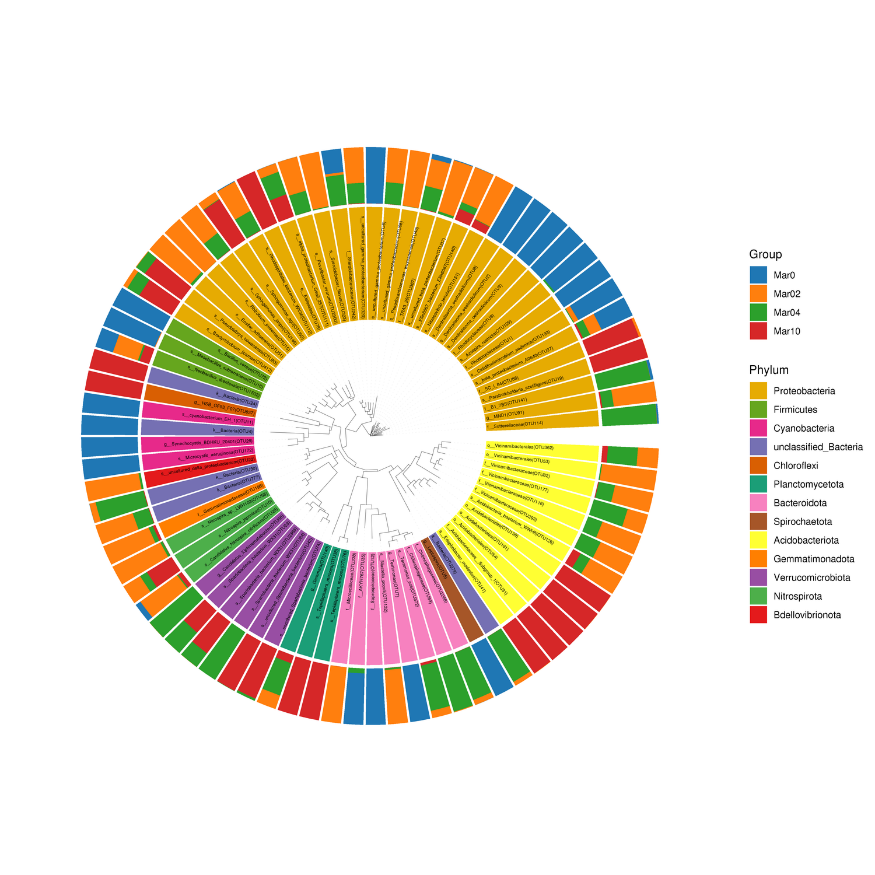
Community distribution phylogenetic tree
Alpha kusiyana kwekuongorora: ACE
Beta siyana kuongororwa: PCoA
Intergroup analysis: ANOVA
Ongorora kufambira mberi kunofambiswa neBMKGene's amplicon sequencing masevhisi nePacBio kuburikidza neakaunganidzwa muunganidzwa wezvinyorwa.
Gao, X. naWang, H. (2023) 'Kuenzanisa Kuongorora kweRumen Bacterial Profiles uye Mabasa panguva yeAdaption to Different Phenology (Regreen vs. Grassy) muAlpine Merino Sheep ine Matanho maviri Anokura paAlpine Meadow', Fermentation, 9( 1), p. 16. doi: 10.3390/FERMENTATION9010016/S1.
Li, S. nevamwe. (2023) 'Kutora microbial yakasviba nyaya muvhu regwenga uchishandisa culturomics-based metagenomics uye high-resolution analysis', npj Biofilms uye Microbiomes 2023 9: 1, 9 (1), mapeji 1-14. doi: 10.1038/s41522-023-00439-8.
Mu, L. et al. (2022) 'Migumo yemafuta aneasidhi munyu pamaitiro ekuvirisa, kusiyana kwebhakitiriya uye kugadzikana kweaerobic yemasilaji akasanganiswa anogadzirwa nealfa, mashanga emupunga uye bran yegorosi', Journal of the Science of Food and Agriculture, 102(4), pp. 1475– 1487. doi: 10.1002/JSFA.11482.
Yang, J. nevamwe. (2023) 'Kudyidzana pakati peOxidative Stress Biomarkers uye Gut Microbiota muAntioxidant Effects yeZvitorwa kubva kuSonchus brachyotus DC. muOxazolone-Induced Intestinal Oxidative Stress in Adult Zebrafish', Antioxidants 2023, Vol. 12, Peji 192, 12(1), p. 192. doi: 10.3390/ANTIOX12010192.