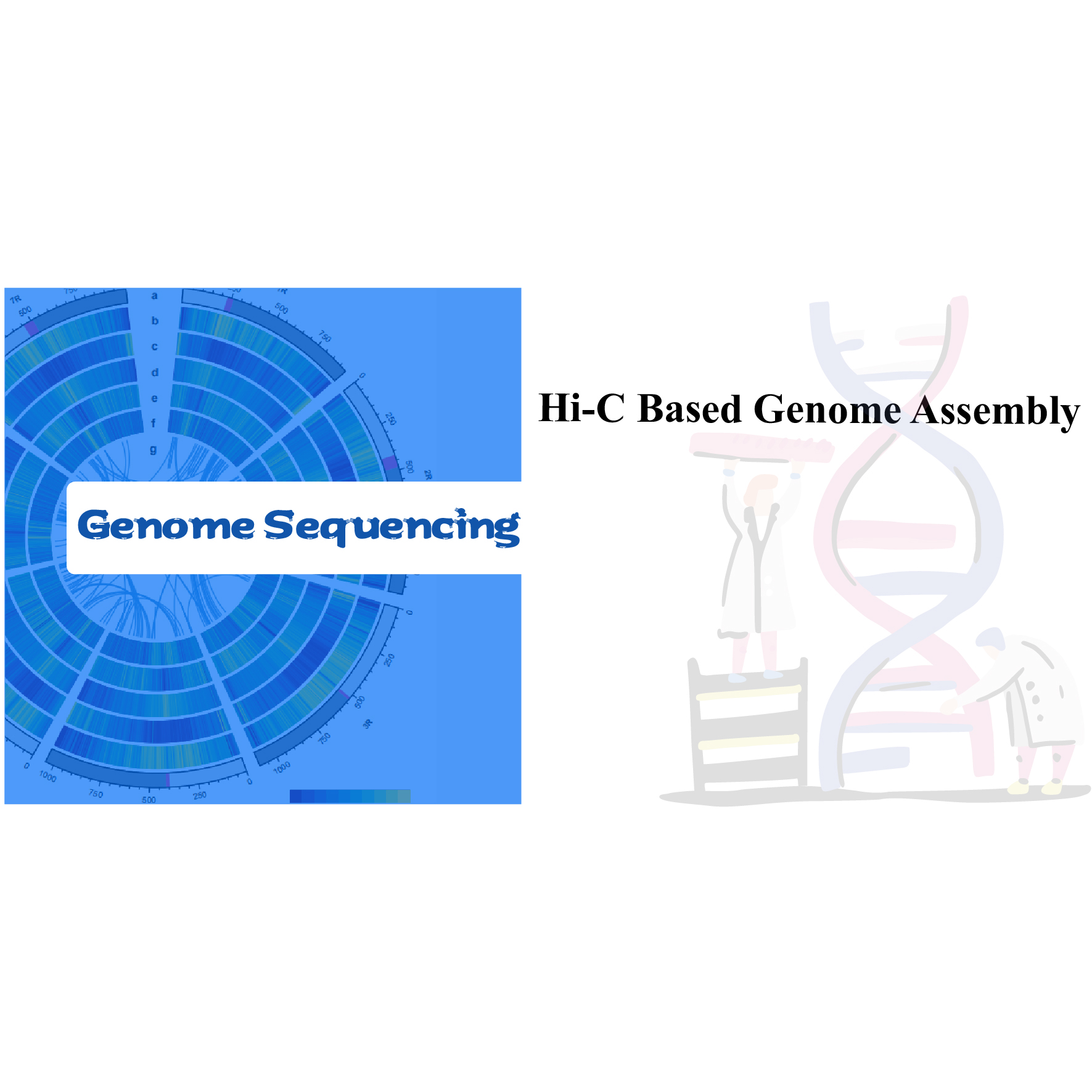
Plant/Animal Whole Genome Sequencing
Service Features
De novo
This technique is particularly useful with challenging genomes such as those with high degree of heterozygosity, repetitive regions, polypoloid genomes, abnormal CG contents etc.
Our one-stop solution provides integrated sequencing services and bioinformatic analysis that deliver a high-quality de novo assembled genome. An initial genome survey with Illumina provides estimations of genome size and complexity, and this information is used to guide the next step of long-read sequencing with PacBio HiFi, followed by de novo assembly of contigs. The subsequent use of HiC assembly enables anchoring of the contigs to the genome, obtaining a chromosome-level assembly. Finally, the genome is annotated by gene prediction and by sequencing expressed genes, resorting to transcriptomes with short and long reads.
-- Integration of multiple sequencing and bioinformatic services in a one-stop solutionService suitable for the constructing novel
-- genomes or improvement of existing reference genomes for species of interest.
Resequencing
-- Library preparation can be standard or PCR-free
-- Available in 4 sequencing platforms: Illumina NovaSeq, MGI T7, Nanopore Promethion P48 or PacBio Revio.
-- Bioinformatic analysis focused on variant calling: SNP, InDel, SV and CNV
Service Advantages
● Extensive Expertise and Publication Record: For de novo services, we have accumulated massive experience in high-quality genome assembly of diverse species, including diploid genomes and highly complex genomes of polyploid and allopolyploid species. Since 2018, we have contributed to over 300 high-impact publications, and 20+ of them are published in Nature Genetics. On genome re-sequecning, we accumulated over 1000 species, which resulted in over 1000 published cases with a cumulative impact factor of over 5000.
● One-stop Solution: On de novo sequencing, our integrated approach combines multiple sequencing technologies and bioinformatic analyses into a cohesive workflow, delivering a high-quality assembled genome.
● Tailored to Your Needs: Our service workflow is customizable, allowing adaptation for genomes with diverse features and specific research needs.
● Highly Skilled Bioinformatics and Laboratorial Team: Whether it is for de novo sequencing or re-sequencing, our team has a skilled set of tools and knowledge to guarantee the project success. This can be corroborated with the series of patents and software copyrights they’ve developed.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
● Comprehensive Bioinformatics Analysis: Including variation calling and function annotation.
● Comprehensive Annotation for Sequencing: We use multiple databases to functionally annotate the genes with identified variations and perform the corresponding enrichment analysis, providing insights on your research projects.
Service Specifications
|
Variants to be identified |
Sequencing strategy |
Recommended depth |
|
SNP and InDel |
Illumina NovaSeq PE150 or MGI T7 |
10x |
|
SV and CNV (less accurate) |
30x |
|
|
SV and CNV (more accurate) |
Nanopore Prom P48 |
20x |
|
SNPs, Indels, SV and CNV |
PacBio Revio |
10x |
Sample Requirements
|
Tissue or extracted nucleic acids |
Illumina/MGI |
Nanopore |
PacBio
|
||
|
Animal Viscera |
0.5-1 g |
≥ 3.5 g
|
≥ 3.5 g
|
||
|
Animal Muscle |
≥ 5 g
|
≥ 5 g
|
|||
|
Mammalian Blood |
1.5 mL |
≥ 0.5 mL
|
≥ 5 mL
|
||
|
Poultry/Fish Blood |
≥ 0.1 mL
|
≥ 0.5 mL
|
|||
|
Plant- Fresh Leaf |
1-2 g |
≥ 2 g
|
≥ 5 g
|
||
|
Cultured Cells |
|
≥ 1x107
|
≥ 1x108
|
||
|
Insect soft tissue/Individual |
0.5-1 g |
≥ 1 g
|
≥ 3 g
|
||
|
Extracted DNA
|
Concentration: ≥ 1 ng/ µL Amount: ≥ 30 ng Limited or no degradation or contamination
|
Concentration Amount
OD260/280
OD260/230
Limited or no degradation or contamination
|
≥ 40 ng/ µL 4 µg/flow cell/sample
1.7-2.2
≥1.5 |
Concentration Amount
OD260/280
OD260/230
Limited or no degradation or contamination |
≥ 50 ng/ µL
10 µg/flow cell/sample
1.7-2.2
1.8-2.5 |
|
PCR-free Library Preparation: Concentration≥ 40 ng/ µL Amount≥ 500 ng |
|||||
Service Work Flow

Sample delivery

DNA extraction

Library construction

Sequencing

Data analysis
Data delivery
De novo bioinformatics pipeline
If you want to see an overview of our different pipelines for the assembly:
Complete bioinformatic analysis, separated in 4 steps:
1. Genome survey, based on k-mer analysis with NGS reads. It will give us information on:
- Estimation of genome size
- Estimation of heterozygosity
- Estimate of repetitive regions
2. Genome Assembly with PacBio HiFi. Using long reads will give us:
- De novo assembly
- Assembly assessment: including BUSCO analysis for genome completeness and mapping back of NGS and PacBio HiFi reads
3. Hi-C assembly. Once we have the assembly, having information on the genome 3D structure will deepen the knowledge and enrich the reference genome we are building.
- Hi-C library QC to estimate valid Hi-C interactions.
- Hi-C assembly. We will do the clustering of contigs in groups, followed by contig ordering within each group and assigning contig orientation.
- Hi-C evaluation
4. Genome annotation:
- Non-coding RNA prediction
- Repetitive sequences identification (transposons and tandem repeats)
- Gene prediction
- De novo: ab initio algorithms
- Based on homology
- Based on transcriptome, with long and short reads: reads are de novo assembled or mapped to the draft genome
- Annotation of predicted genes with multiple databases
Re-sequencing bioinformatics pipeline
Includes the following analysis:
- Raw Data Quality Control
- Statistics of alignment to reference genome
- Variant identification: SNP, InDel, SV and CNV
- Functional annotation of variants
Statistics of alignment to reference genome – sequencing depth distribution
SNP calling among multiple samples
InDel identification – statistics of the InDel length in the CDS region and the genome-wide region
Variant distribution across the genome – Circos plot
Functional annotation of genes with identified variants – Gene Ontology
Chai, Q. et al. (2023) ‘A glutathione S‐transferase GhTT19 determines flower petal pigmentation via regulating anthocyanin accumulation in cotton’, Plant Biotechnology Journal, 21(2), p. 433. doi: 10.1111/PBI.13965.
Cheng, H. et al. (2023) ‘Chromosome-level wild Hevea brasiliensis genome provides new tools for genomic-assisted breeding and valuable loci to elevate rubber yield’, Plant Biotechnology Journal, 21(5), pp. 1058–1072. doi: 10.1111/PBI.14018.
Li, C. et al. (2021) ‘Genome sequences reveal global dispersal routes and suggest convergent genetic adaptations in seahorse evolution’, Nature Communications, 12(1). doi: 10.1038/S41467-021-21379-X.
Li, Y. et al. (2023) ‘Large-Scale Chromosomal Changes Lead to Genome-Level Expression Alterations, Environmental Adaptation, and Speciation in the Gayal (Bos frontalis)’, Molecular Biology and Evolution, 40(1). doi: 10.1093/MOLBEV/MSAD006.
Tian, T. et al. (2023) ‘Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm’, Nature Genetics 2023 55:3, 55(3), pp. 496–506. doi: 10.1038/s41588-023-01297-y.
Zhang, F. et al. (2023) ‘Revealing evolution of tropane alkaloid biosynthesis by analyzing two genomes in the Solanaceae family’, Nature Communications 2023 14:1, 14(1), pp. 1–18. doi: 10.1038/s41467-023-37133-4.
Zeng, T. et al. (2022) ‘Analysis of genome and methylation changes in Chinese indigenous chickens over time provides insight into species conservation’, Communications Biology, 5(1), pp. 1–12. doi: 10.1038/s42003-022-03907-7.
Challenging case-studies:
Telomere-to-telomere assembly: Fu, A. et al. (2023) ‘Telomere-to-telomere genome assembly of bitter melon (Momordica charantia L. var. abbreviata Ser.) reveals fruit development, composition and ripening genetic characteristics’, Horticulture Research, 10(1). doi: 10.1093/HR/UHAC228.
Haplotype assembly: Hu, W. et al. (2021) ‘Allele-defined genome reveals biallelic differentiation during cassava evolution’, Molecular Plant, 14(6), pp. 851–854. doi: 10.1016/j.molp.2021.04.009.
Giant genome assembly: Yuan, J. et al. (2022) ‘Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii’, Nature Communications 2022 13:1, 13(1), pp. 1–16. doi: 10.1038/s41467-022-35063-1.
Polyploid genome assembly: Zhang, Q. et al. (2022) ‘Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum’, Nature Genetics 2022 54:6, 54(6), pp. 885–896. doi: 10.1038/s41588-022-01084-1.