Long Non-coding Sequencing-Illumina
Service Advantages
● Joint analysis of mRNA and lncRNA: by combining the quantification of mRNA transcripts with the study of lncRNA and their targets, it is possible to get an in-depth overview of the regulatory mechanism underlying the cellular response.
● Extensive Expertise: We have processed over 230,000 samples, spanning diverse sample and projects’ objectives. We bring the wealth of expertise to every project.
● Joint analysis of mRNA and lncRNA: We combine the quantification of mRNA transcripts with the study of lncRNA and their targets, making it possible to get an in-depth overview of the regulatory mechanism underlying the cellular response.
● Rigorous Quality Control: We implement core control points across all stages, from sample preparation to library preparation, sequencing and bioinformatics. Our meticulous monitoring ensures the delivery of consistently high-quality results.
● Comprehensive Annotation: We use multiple databases to functionally annotate the Differentially Expressed Genes (DEGs) and perform corresponding enrichment analyses. This comprehensive approach provides insights into the cellular and molecular processes underlying the transcriptome response, ensuring you get all the information possible about your experiment’s data.
● Post-Sales Support: We understand how important is to be present, that’s why our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Sample Requirements and Delivery
|
Library |
Platform |
Recommended data |
Data QC |
|
rRNA depleted directional library |
Illumina PE150 |
10-16 Gb |
Q30≥85% |
Nucleotides:
|
Conc.(ng/μl) |
Amount (μg) |
Purity |
Integrity |
|
≥ 80 |
≥ 0.8 |
OD260/280=1.7-2.5 OD260/230=0.5-2.5 Limited or no protein or DNA contamination shown on gel. |
RIN≥6.0; 5.0≥28S/18S≥1.0; limited or no baseline elevation |
● Plants:
Root, Stem or Petal: 450 mg
Leaf or Seed: 300 mg
Fruit: 1.2 g
● Animal:
Heart or Intestine: 450 mg
Viscera or Brain: 240 mg
Muscle: 600 mg
Bones, Hair or Skin: 1.5g
● Arthropods:
Insects: 9g
Crustacea: 450 mg
● Whole blood: 2 tubes
● Cells: 106 cells
● Serum and Plasma: 6 mL
Recommended Sample Delivery
Container: 2 ml centrifuge tube (Tin foil is not recommended)
Sample labeling: Group+replicate e.g. A1, A2, A3; B1, B2, B3.
Shipment:
1. Dry-ice: Samples need to be packed in bags and buried in dry-ice.
2. RNAstable tubes: RNA samples can be dried in RNA stabilization tube(e.g. RNAstable®) and shipped in room temperature.
Service Work Flow

Experiment design

Sample delivery

RNA extraction

Library construction

Sequencing

Data analysis

After-sale services
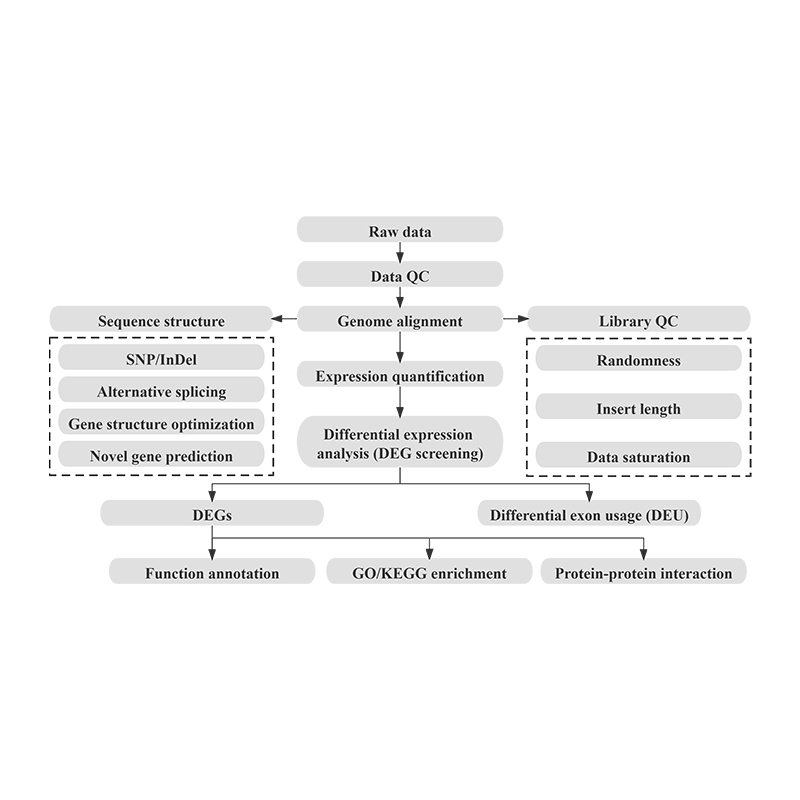
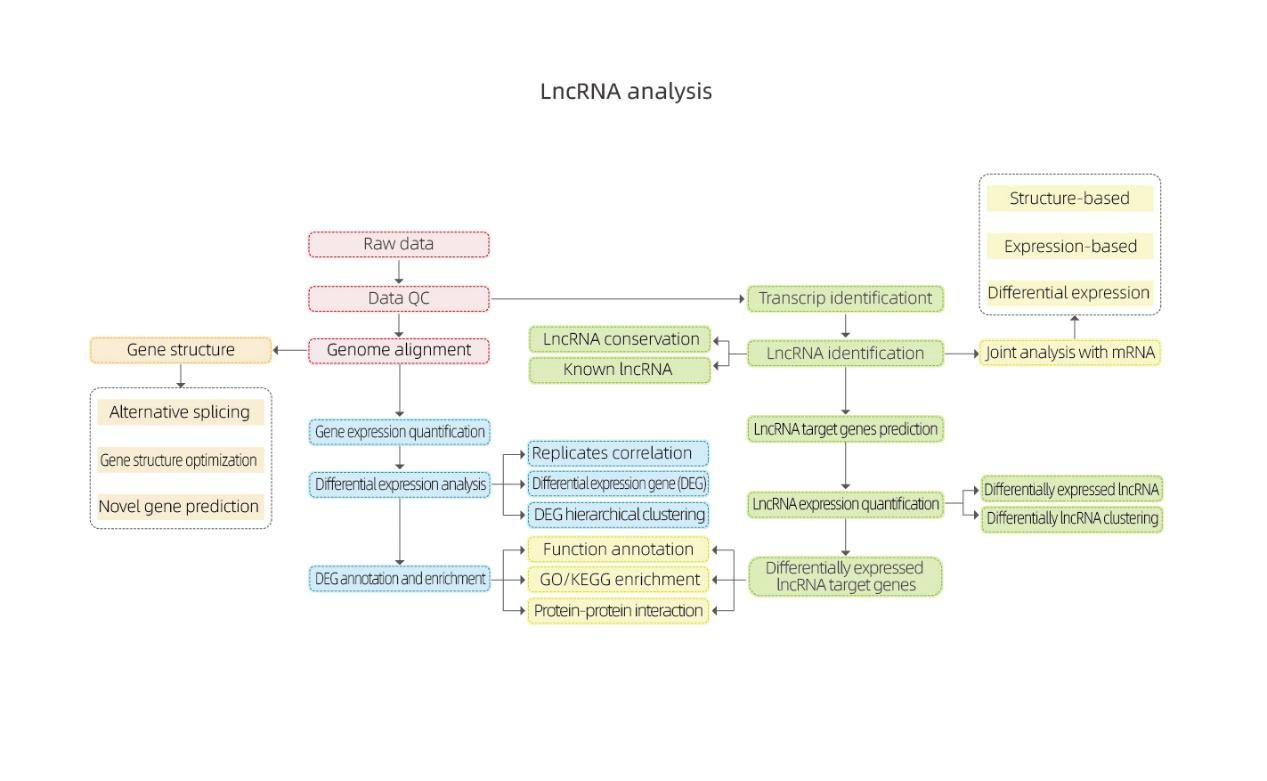
Bioinformatics
- Raw data
- Data QC
- Genome alignment
- Gene structure (Alternative splicing, gene structure optimization and novel gene prediction)
- Gene expression quantification
- Differential expression analysis
- DEG annotation and enrichment + Differential expressed lncRNA target genes
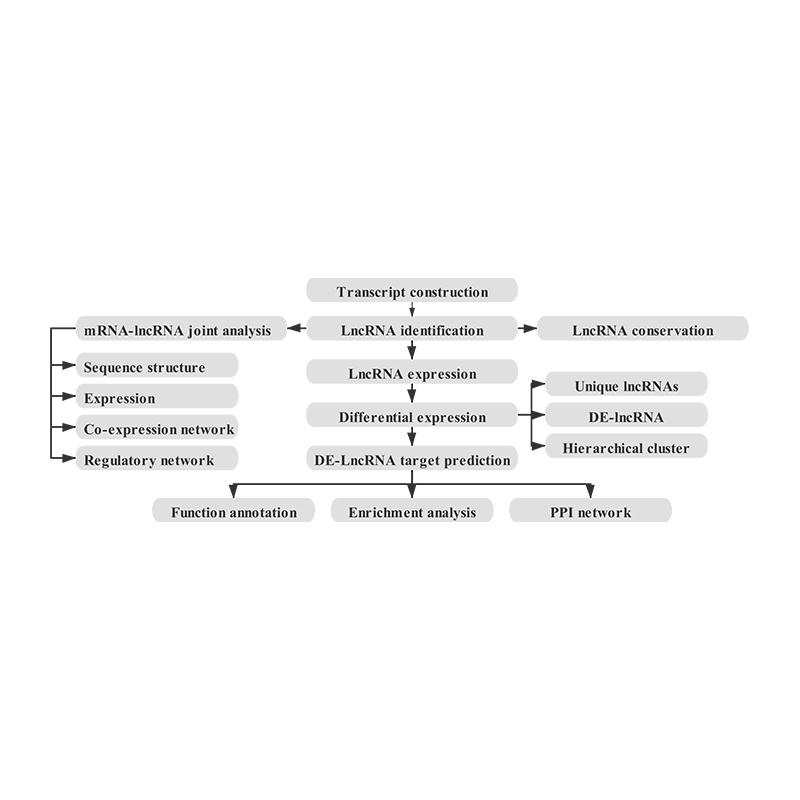
- Transcript identification
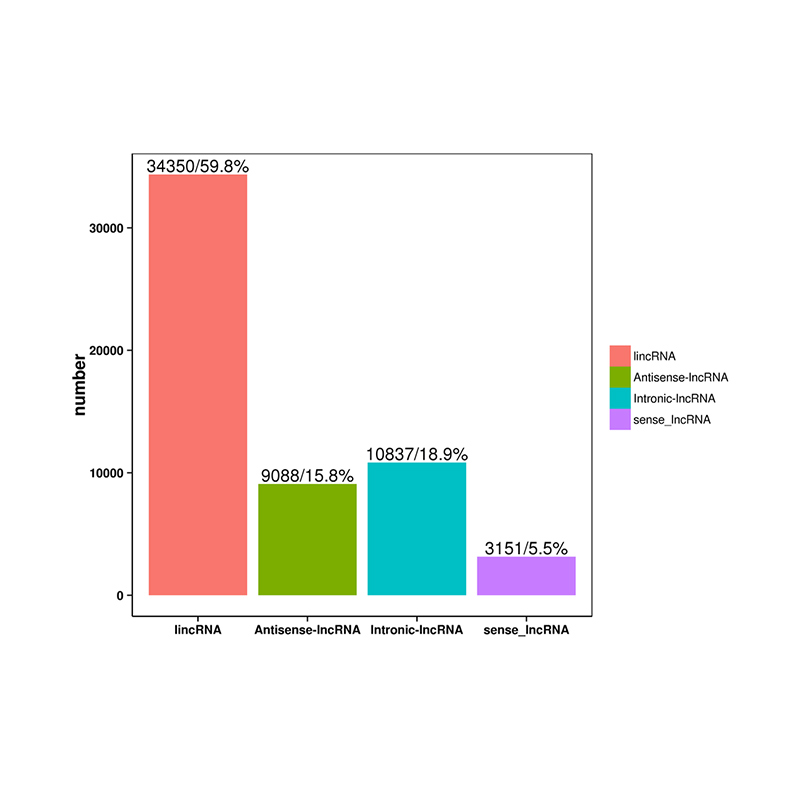
- lncRNA identification (lncRNA conservation and known lncRNA)
- lncRNA target genes prediction
- lncRNA expression quantification
- Joint analysis with mRNA data
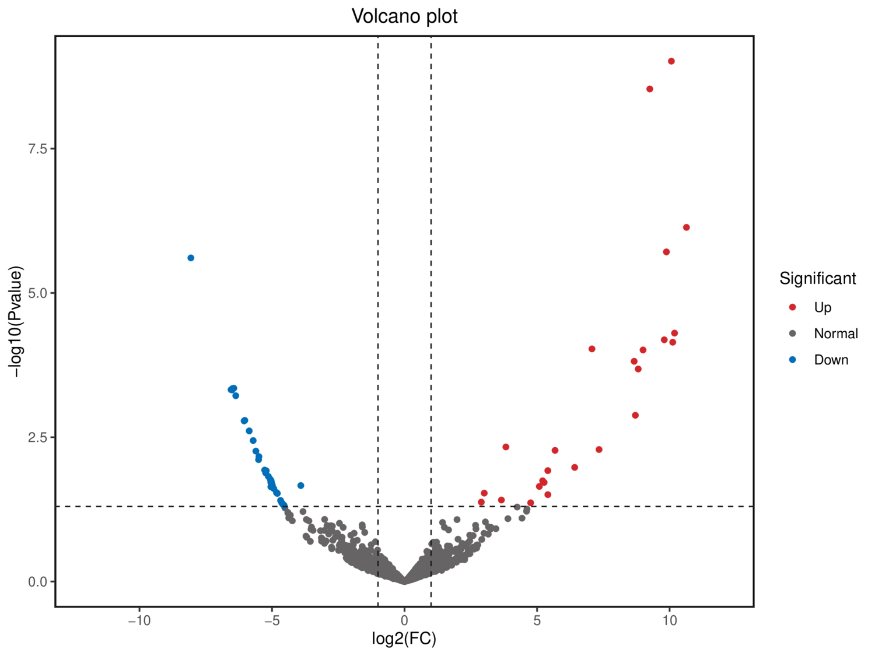
Differential Gene Expression (DEGs) analysis
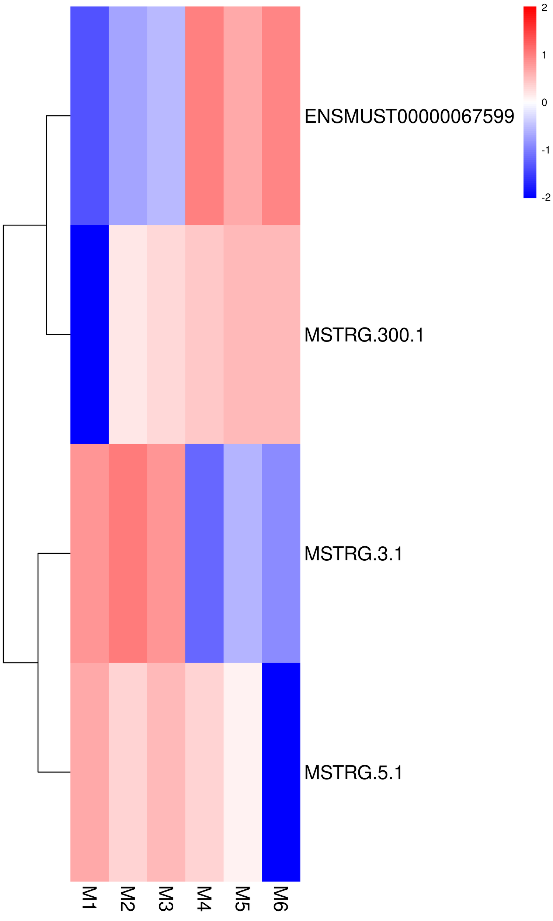
Quantification of lncRNA expression – clustering
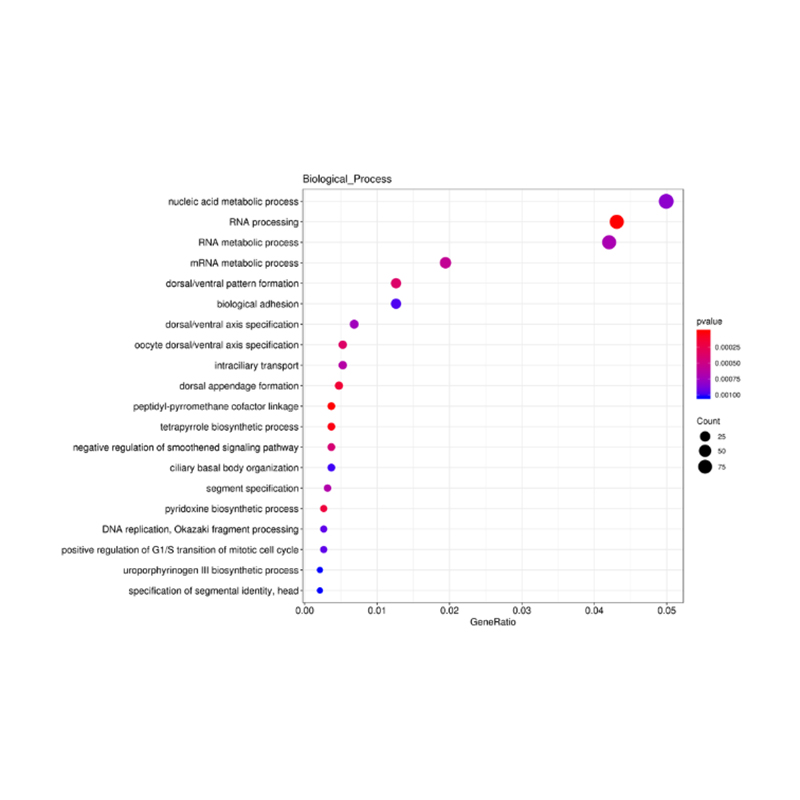
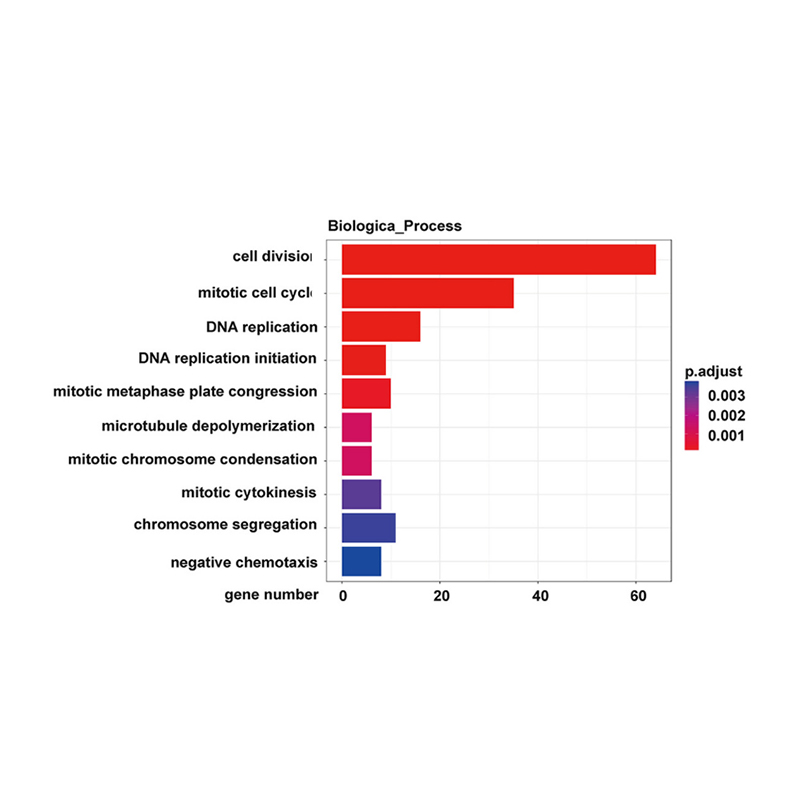
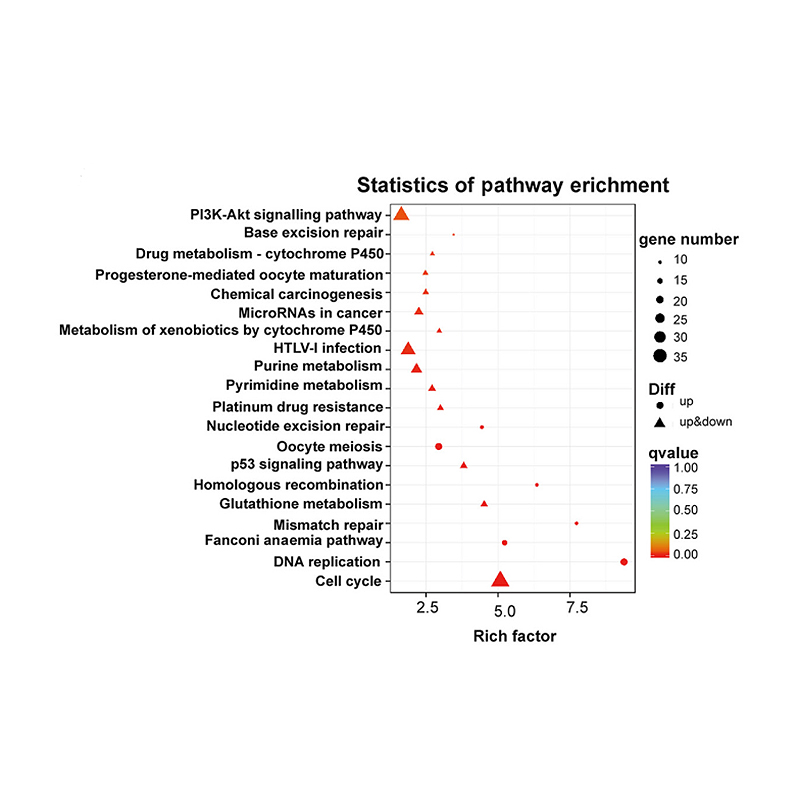
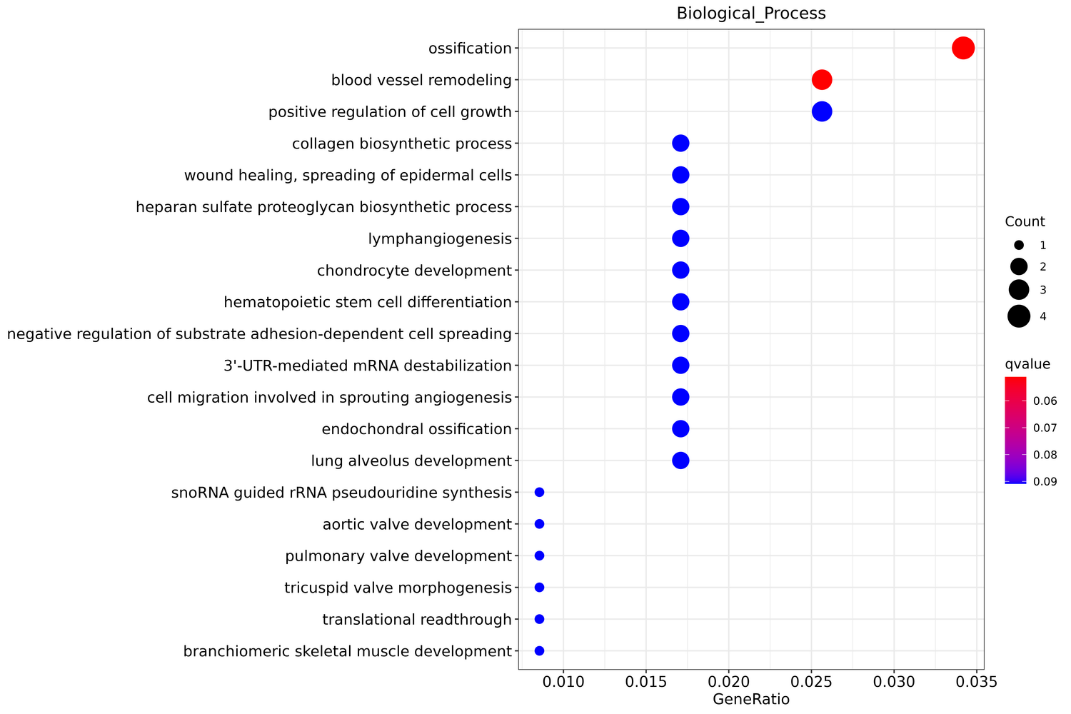
Enrichment of lncRNA target genes
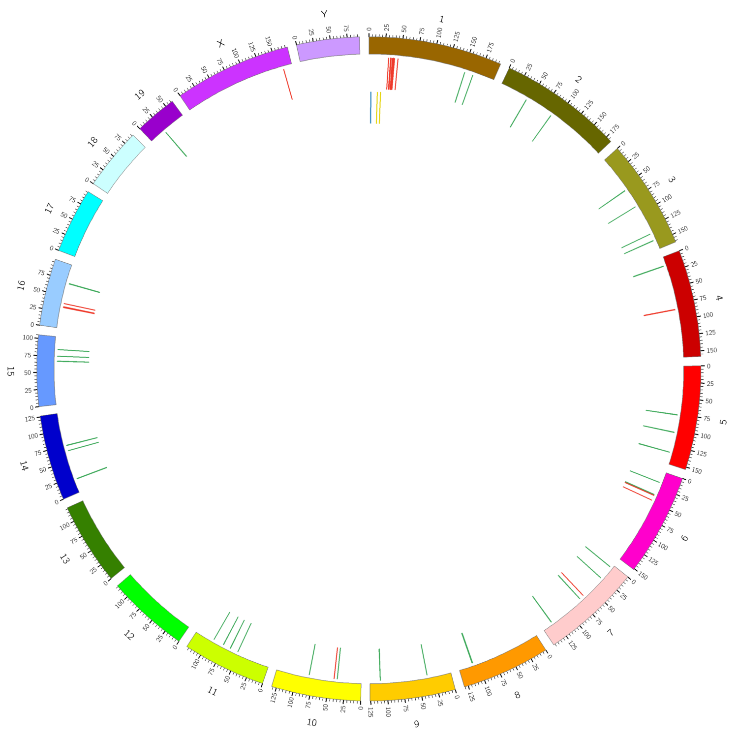
Joint mRNA and lncRNA position analysis – Circos plot (middle circle is mRNA and inner cicrlce is lncRNA)
Explore the advancements facilitated by BMKGene’s lncRNA equencing services through a curated collection of publications.
Ji, H. et al. (2020) ‘Identification, functional prediction, and key lncRNA verification of cold stress-related lncRNAs in rats liver’, Scientific Reports 2020 10:1, 10(1), pp. 1–14. doi: 10.1038/s41598-020-57451-7.
Jia, Z. et al. (2021) ‘Integrative Transcriptomic Analysis Reveals the Immune Mechanism for a CyHV-3-Resistant Common Carp Strain’, Frontiers in Immunology, 12, p. 687151. doi: 10.3389/FIMMU.2021.687151/BIBTEX.
Wang, X. J. et al. (2022) ‘Multi-Omics Integration-Based Prioritisation of Competing Endogenous RNA Regulation Networks in Small Cell Lung Cancer: Molecular Characteristics and Drug Candidates’, Frontiers in Oncology, 12, p. 904865. doi: 10.3389/FONC.2022.904865/BIBTEX.
Xiao, L. et al. (2020) ‘Genetic dissection of the gene coexpression network underlying photosynthesis in Populus’, Plant Biotechnology Journal, 18(4), pp. 1015–1026. doi: 10.1111/PBI.13270.
Zheng, H. et al. (2022) ‘A Global Regulatory Network for Dysregulated Gene Expression and Abnormal Metabolic Signaling in Immune Cells in the Microenvironment of Graves’ Disease and Hashimoto’s Thyroiditis’, Frontiers in Immunology, 13, p. 879824. doi: 10.3389/FIMMU.2022.879824/BIBTEX.