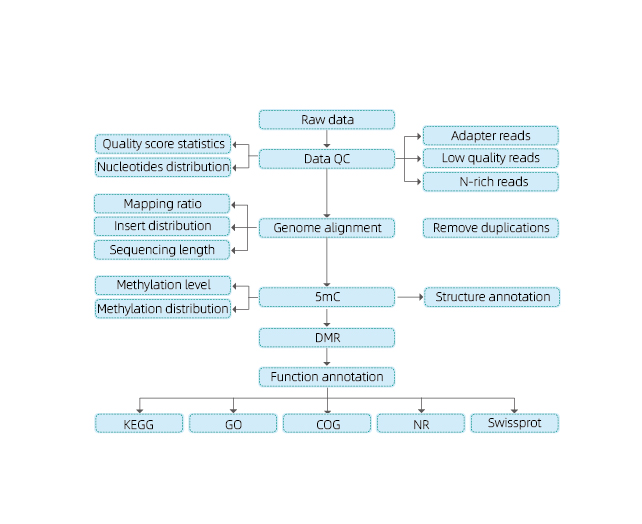
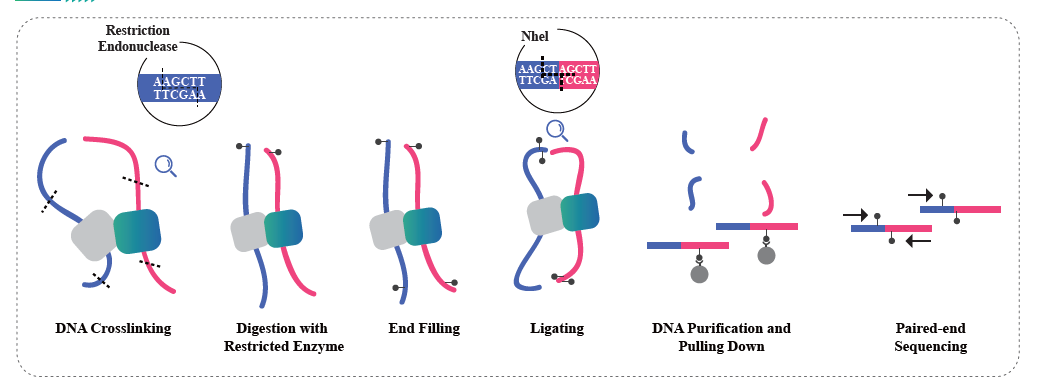
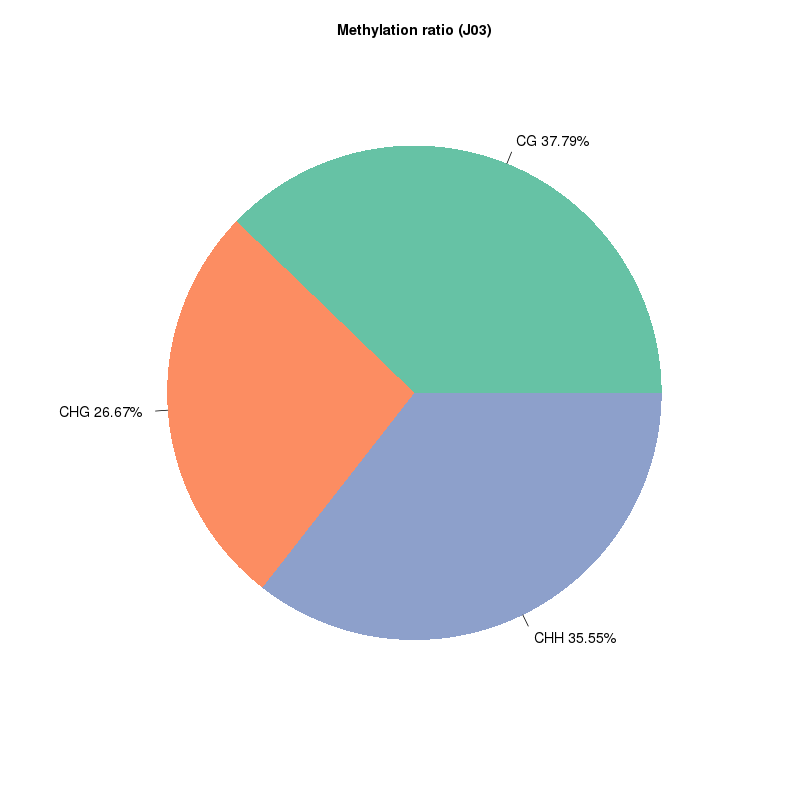Totum genome bisul fi te sequencing (wgbs)
Service Features
● requirit referatur.
● Lambda DNA est additum ad Monitor Bisulfite conversionem efficientiam.
● sequencing in illumina novaseq.
Service commoda
●Aurum Latin quia DNA Methylation Research: Haec Matura Methylation Conversion Processing Technology habet princeps accurate et bonum reproducibility.
●Wide coverage et uno-basi resolutio:Deprehensio de methylation sites ad genome, wide gradu.
●Complete Platform:Providere unum-subsisto optimum servitium a sample dispensando, bibliotheca constructione, sequencing ad bioinformatics analysis.
●Extensive expertiseIn WGBs sequencing projects feliciter perficitur per diversum rhoncus speciei, BMKGEGGE in a decade de experientia, a valde peritus analysis quadrigis, comprehensive contentus, et excellens post-Sales auxilium.
●Facultatem ad iungere et transcripomomics analysis: Permittens in integrated analysis WGBs cum aliis omics data ut RNA, seq.
Sample specifications
| Bibliotheca | Sequencing belli | Commendatur data output | Imperium |
| Tractatus | Illumina pe150 | 30x profundum | Q30 ≥ LXXXV% Bisulfite conversionem> XCIX% |
Sample Requirements
| Concentration (ng / μl) | Summa copia (μg) | Additional requisitis | |
| Genomic dna | ≥ V | ≥ CD NG | Limited degradation aut contaminationem |
Service opus influunt

Sample partum

DNA extraction

Constructio

Sequencing

Analysis
Notitia traditio
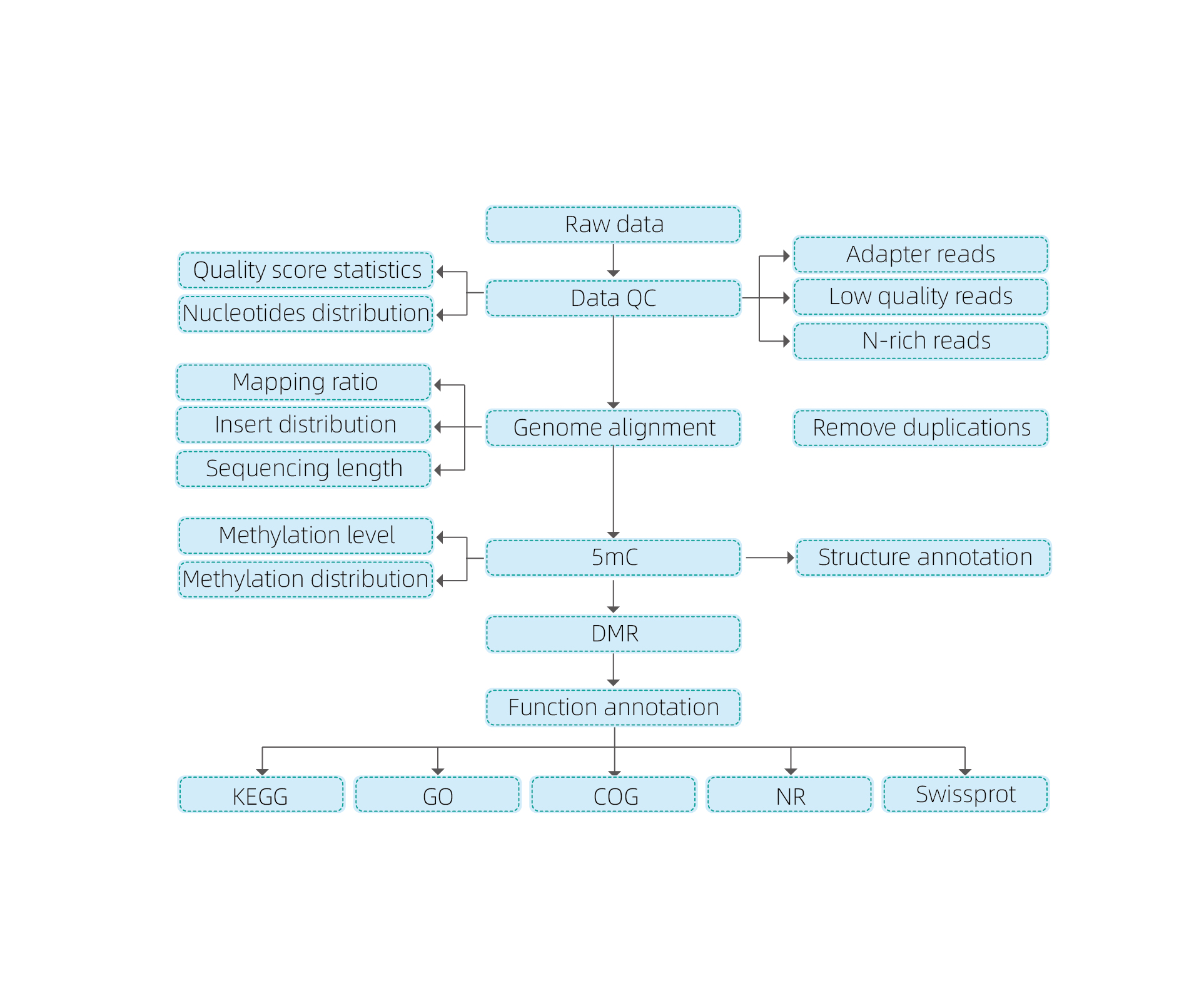
Includit sequenti analysis:
● rudis sequencing qualis imperium;
● Mapping ad reference genome;
● deprehendatur 5mc methylated bases;
● Analysis de methylation distribution et annotation;
● analysis de differentialy methylated regiones (DMRS);
● Functional annotation de genes consociata ad DMRS.
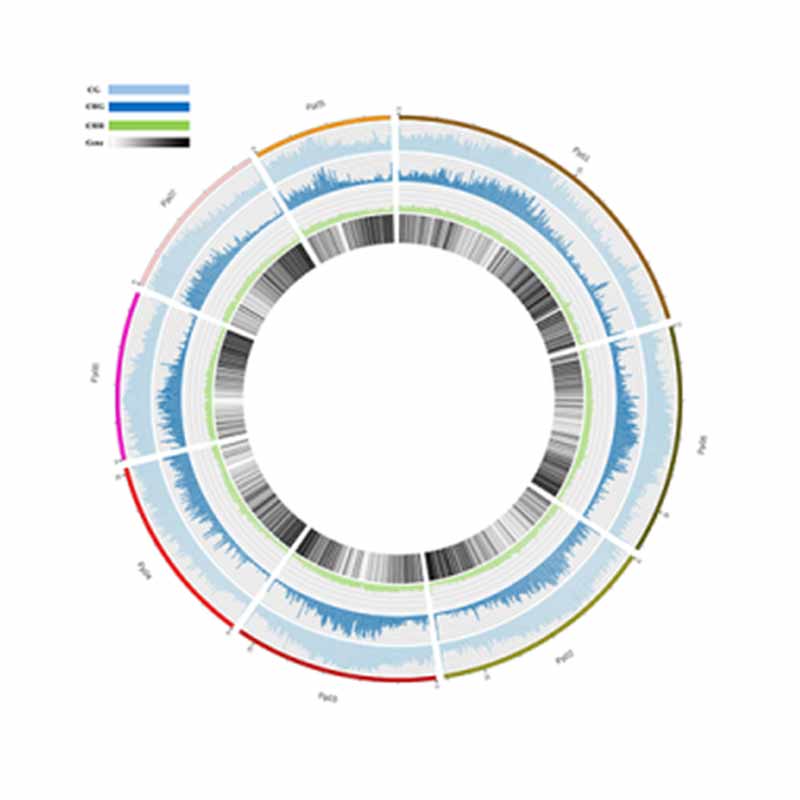
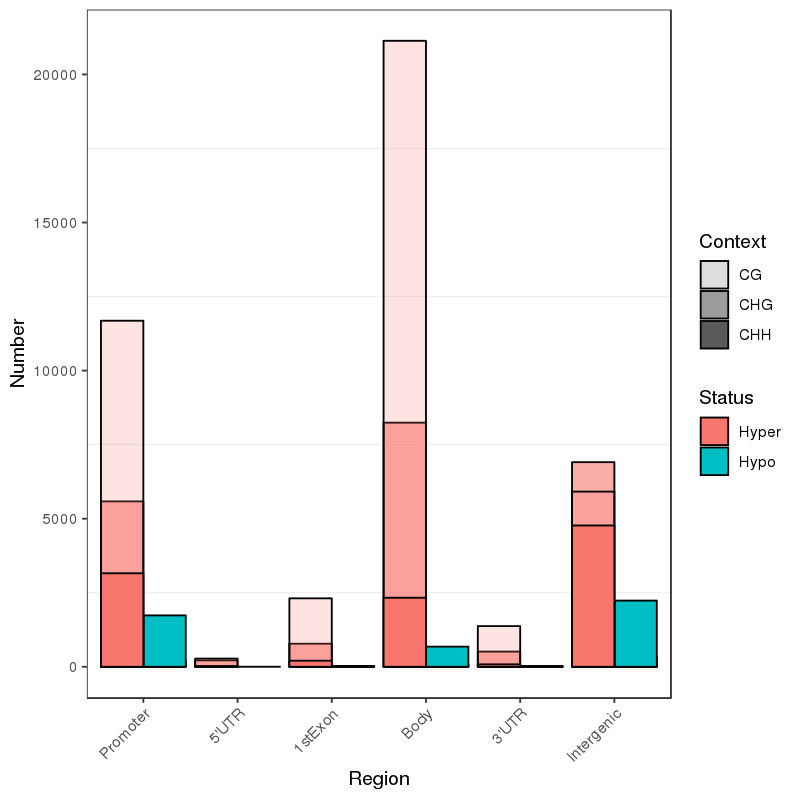
5mc methylation deprehendatur: Genera Methylated Sites
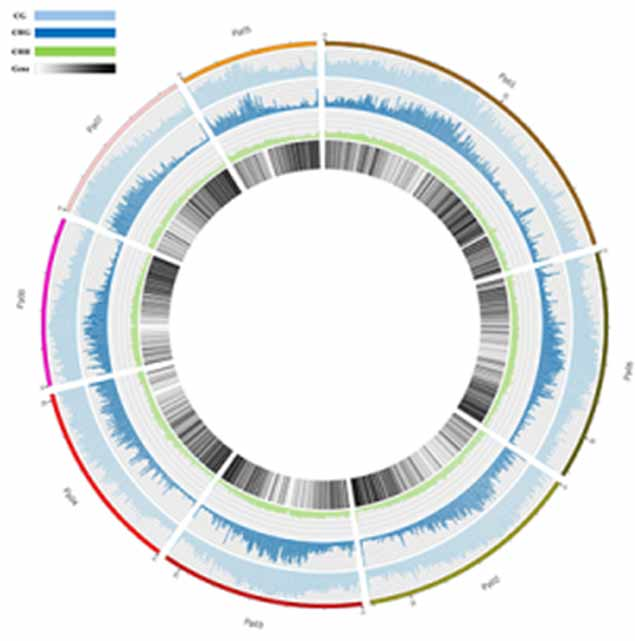
Methylation map. 5mc methylation-wide distribution
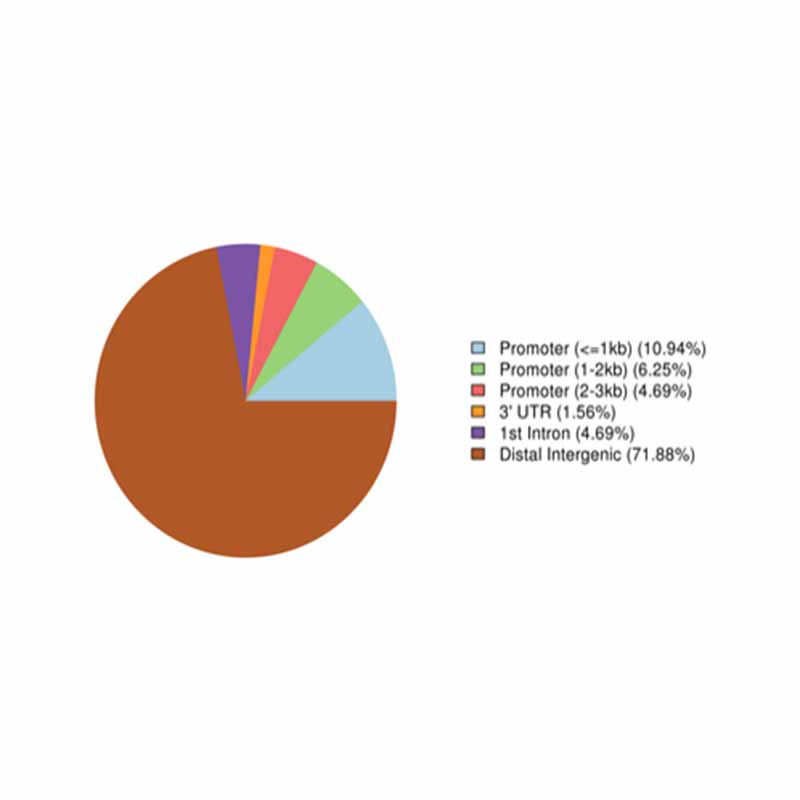
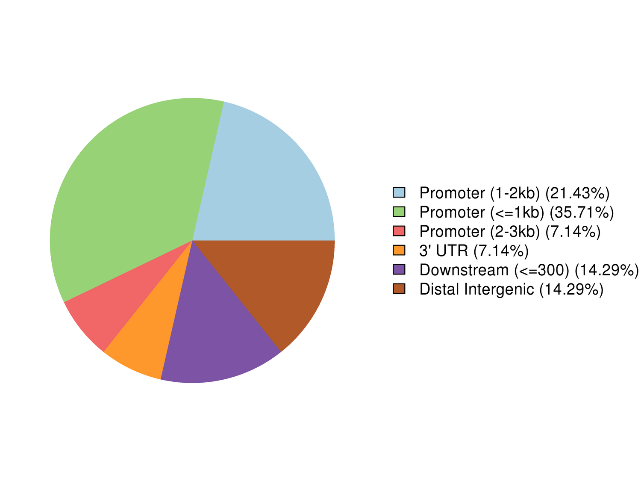
Annotation of altus methylated regiones
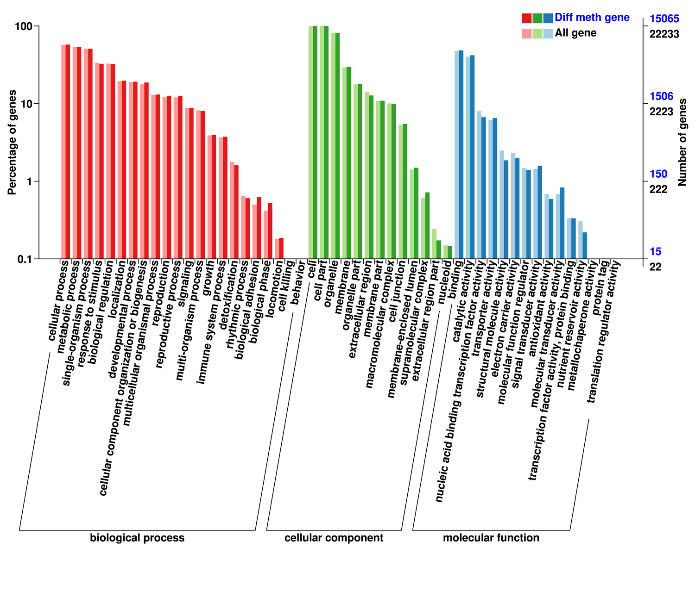
Diverseially Methylated regiones: consociata genes
Diverseially Methylated regiones: Annotation de consociata genes (Gene Ontology)
Explore in investigationis progressiones facilitati per BMKGene est tota genome bisulfite sequencet Services per cated collection of publications.
Fan, Y. et al. (MMXX), Analysis de DNA methylation profiles in ovis Osseus musculus development usura tota-genome bisulfite sequencing,BMC genomics, XXI (I), pp. 1-15. Doi: 10.1186 / S12864-020-6751-5.
Zhao, X. et al. (MMXXII), Novel deoxyribonucleic acidum methylation perturbationes in operarios expositae Vinyl chloride ';Toxicology et industriae salutem, XXXVIII (VII), pp. 377-388. Doi: 10.1177 / 07482337221098600
Zuo, J. et al. (MMXX), relationes inter Genome methylation, campester of non-coding rnas, Mrnas et metabolitarum in maturescere lycopersiciSusceptibility fructus,Et Plant Journal, CIII (III), pp. 980-994. Doi: 10.11111 / Tpj.14778.