SPECIE-Locus Amplified fragmentum sequencing (Slaf, seq)
Service Features
● sequencing in novase cum Pe150.
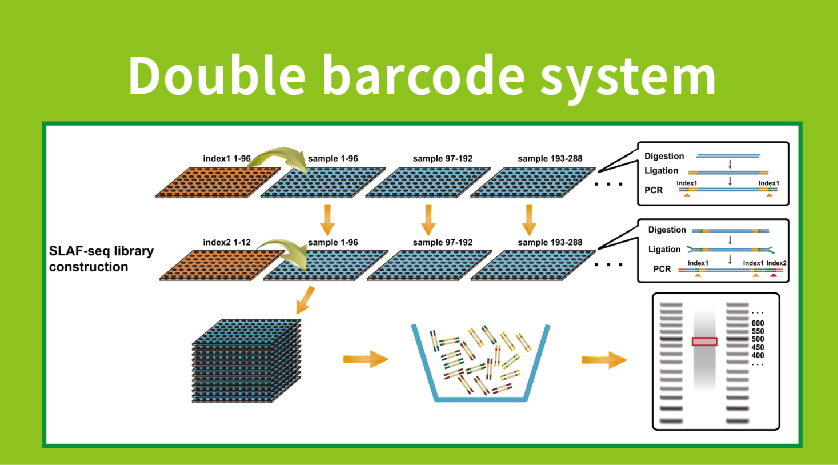
● Library praeparatio cum duplex Barcoding, enabling pooling de super M samples.
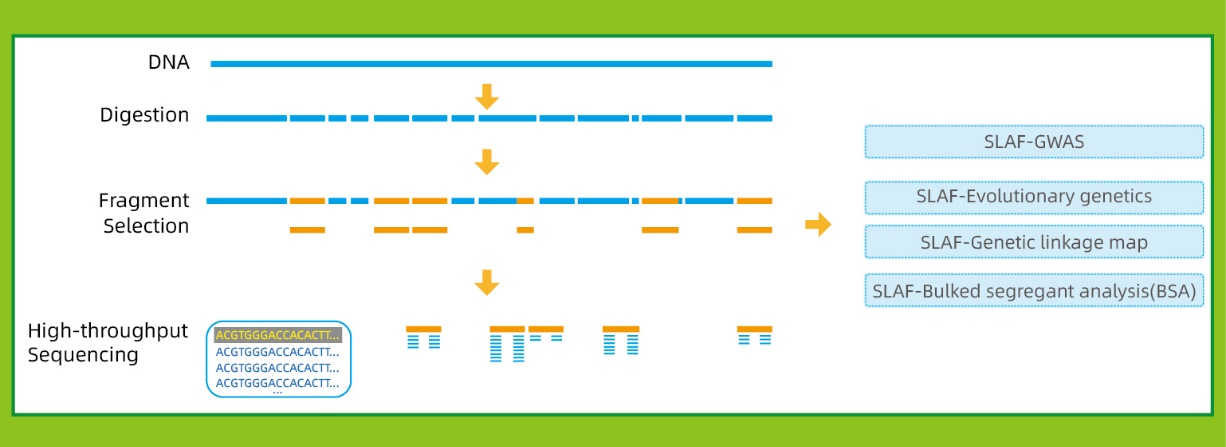
● Hoc ars potest esse cum vel sine referatur Genome, cum diversis bioinformatic Pipelines pro se casu:
Cum reference Genome: SNP et Indel Inventionis
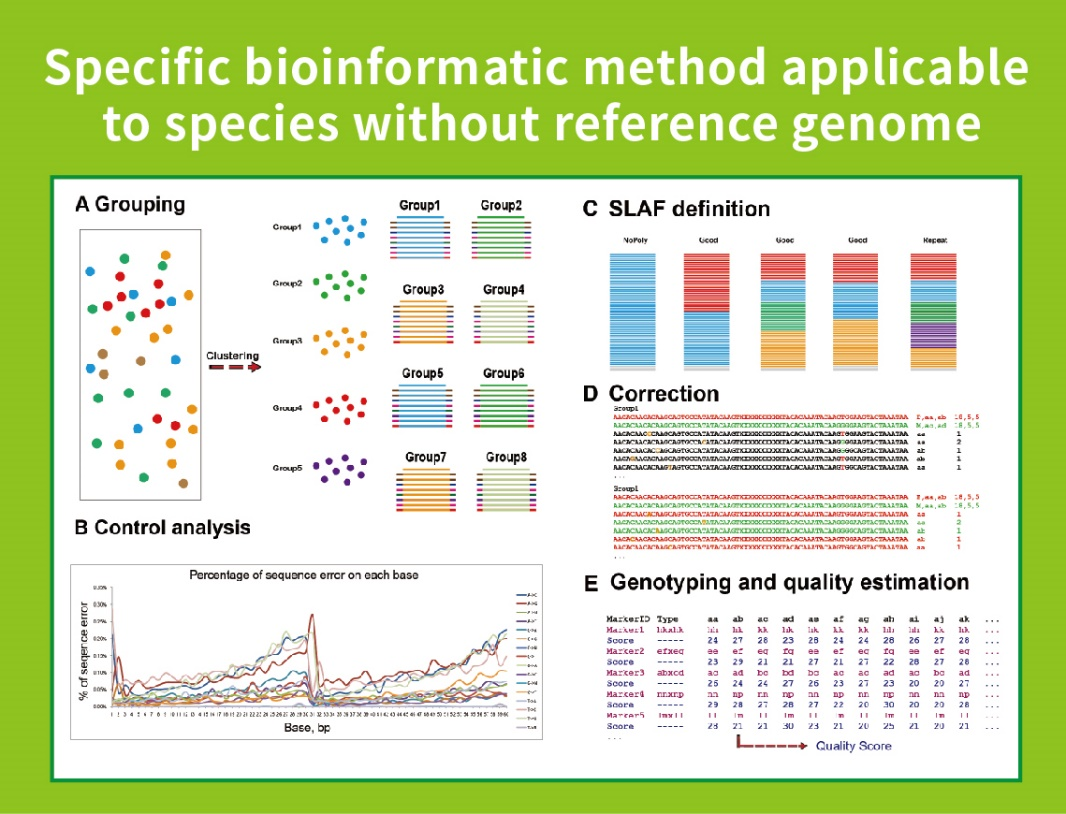
Sine reference Genome: Sample Clustering et SNP Inventionis
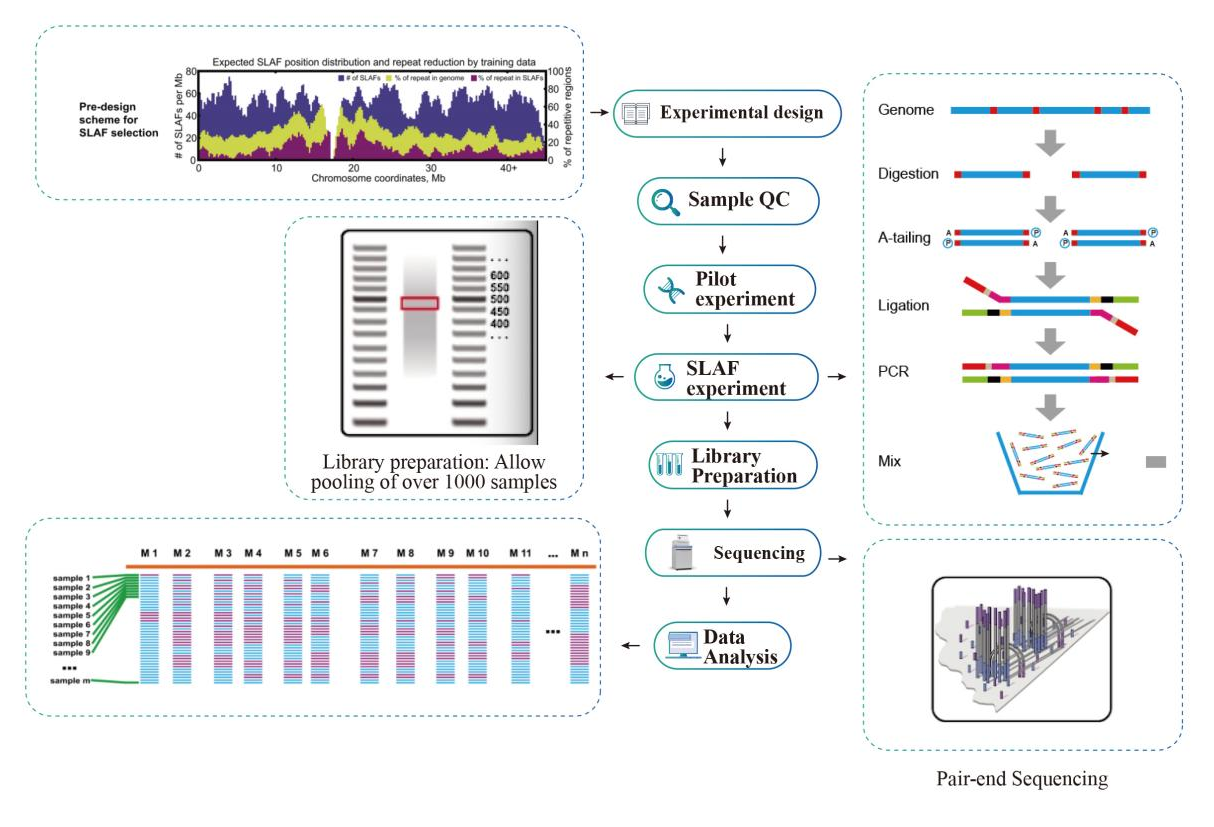
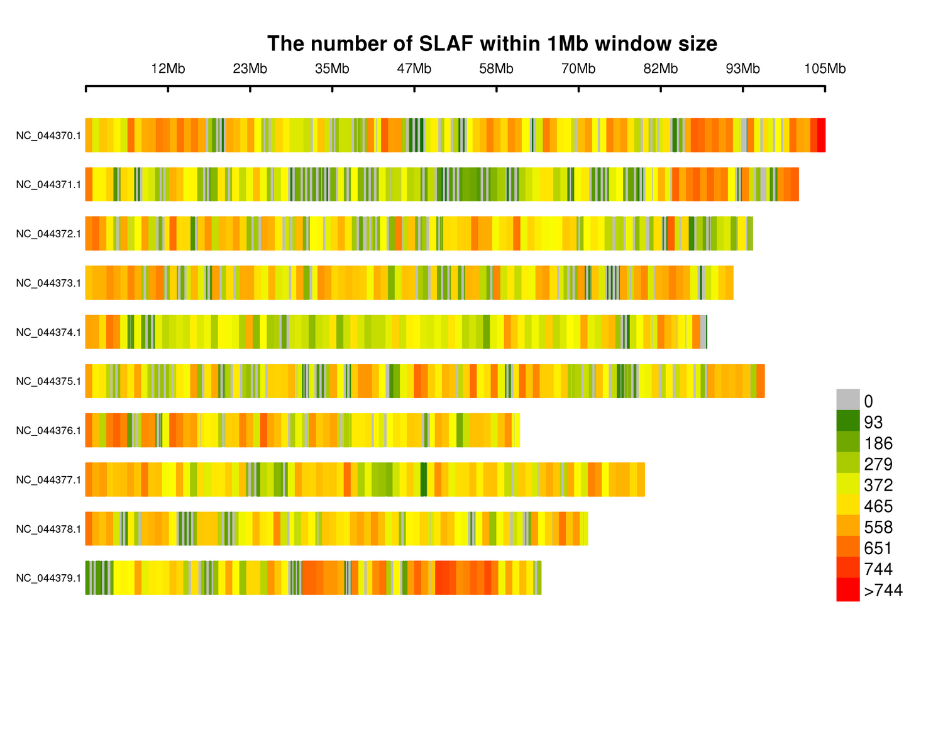
● inin silicoPre-consilio gradu multiple restrictione enzyme junctiones texunt invenire ones, quae generate uniformis distribution de Tags una genome.
● per pre-experimentum, tres enzyme junctiones probata in III exempla ad generate IX Slaf libraries, et hoc notitia est eligere meliorem restrictionem enzyme combination ad project.
Service commoda
●High Genetic Venalicium Inventionis: Integrating a summus throughput duplici barcode ratio concedit ad simultaneous sequencing de magnis populis et locus-specifica amplificationem enhances efficientiam, cursus Tag Numbers occursum diversa requisita variis investigationis quaestionum.
● Minimum dependentia in Genome: Non potest applicari ad speciem cum vel sine referatur Genome.
●Flexibile consilium consilium: Una, enzyme, dual-enzyme, multi, enzyme digestionem, et varia enzymes potest omnes delecti ad opsonationem ad diversum investigationis metas vel speciei. Inin silicoPre-consilio ferri ut optimal enzyme consilio.
● Maximum efficientiam in enzymatica digestio: De conduction estin silicoPre-consilio et pre-experimentum certus optimal consilio cum etiam distribution de Tags in chromosoma (I Slaf Tag / 4kb) et reducitur repetitive sequence (<V%).
●Extensive expertise: Nostra quadrigis affert divitiis experientia omnibus project, cum track record of claudendo super (V) Slaf-seq projects in centum speciei, inter plantis, mammalia, aves, insectorum, et aquatilium organizati.
● Auto-developed bioinformatic workflow: BMKGene developed integrated bioinformatic workflow ad Slaf-seq ut reliability et accurate ad ultima output.
Service Specifications
| Typus of analysis | Commendatur population scale | Sequencing belli | |
| Profundum tag sequencing | Tag numerus | ||
| Geneticae Maps | II parentes et> CL fetus | Parentes: 20x WGS Offsping: 10x | Genome Size: <CD Mb: WGS commendatur <1GB, 100k Tags 1-2GB :: 200K Tags > 2GB, 300k Tags Max 500k Tags |
| Genome, wide Association Studies (Gwas) | ≥200 sample | 10x | |
| Geneticae evolution | ≥30 samples, cum> X exempla ex se subgroup | 10x | |
Service Requirements
Concentration ≥ V NG / μl
Summa moles ≥ LXXX NG
Nanodrop OD260 / CCLXXX = 1.6-2.5
Agarose Gel: Non aut limitata degradation vel contaminationem
Commendatur Sample partus
Continens: II ML Centrifuge TUBULUM
(Plerique exempla, nos suadeo ne conserva in Ethanol)
Sample Labeling: Exempla postulo ut scilicet intitulatum et identical ut summitto sample information forma.
Shipment: siccum-glacies: Exempla postulo ut facis in sacculos primo et sepultus in sicco-glacies.
Opera workflow






Sample QC
PILOTOR
Slaf-experimentum
DECRARATOR
Sequencing
Analysis
Post-Sale Services
 Includit sequenti analysis:
Includit sequenti analysis:
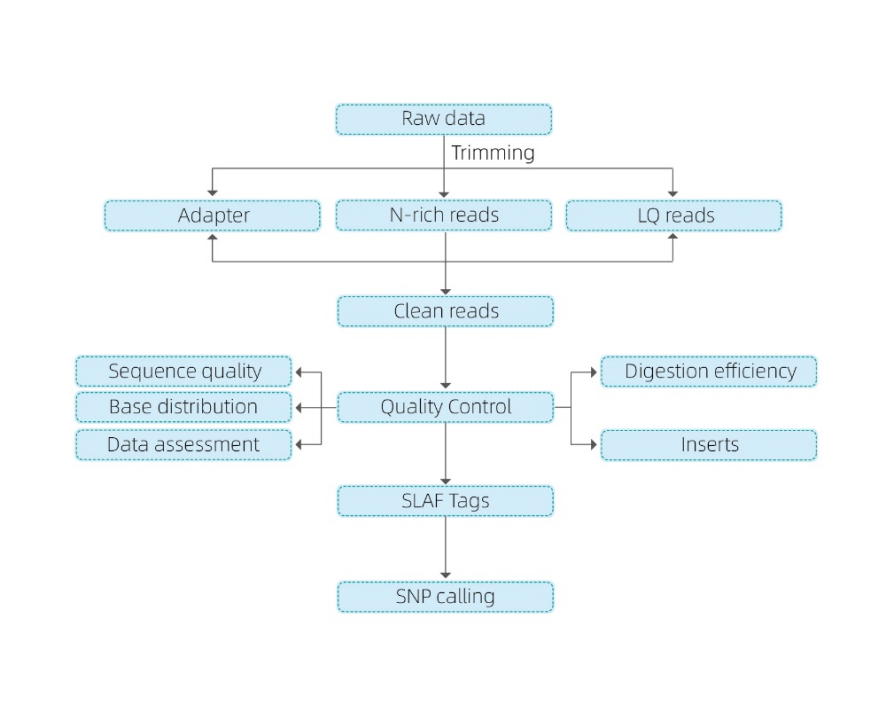
- Sequencing Data QC
- Slaf Tag Development
Mapping ad reference
Sine reference Genome: clustering
- Analysis de Slafe tags.: Statistics, distribution per Genome
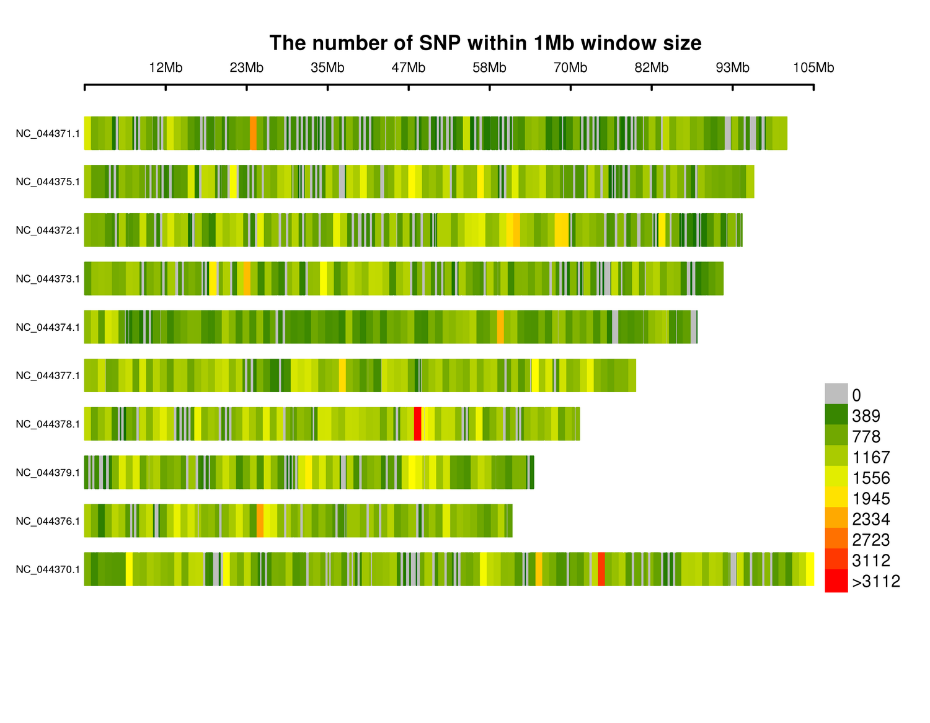
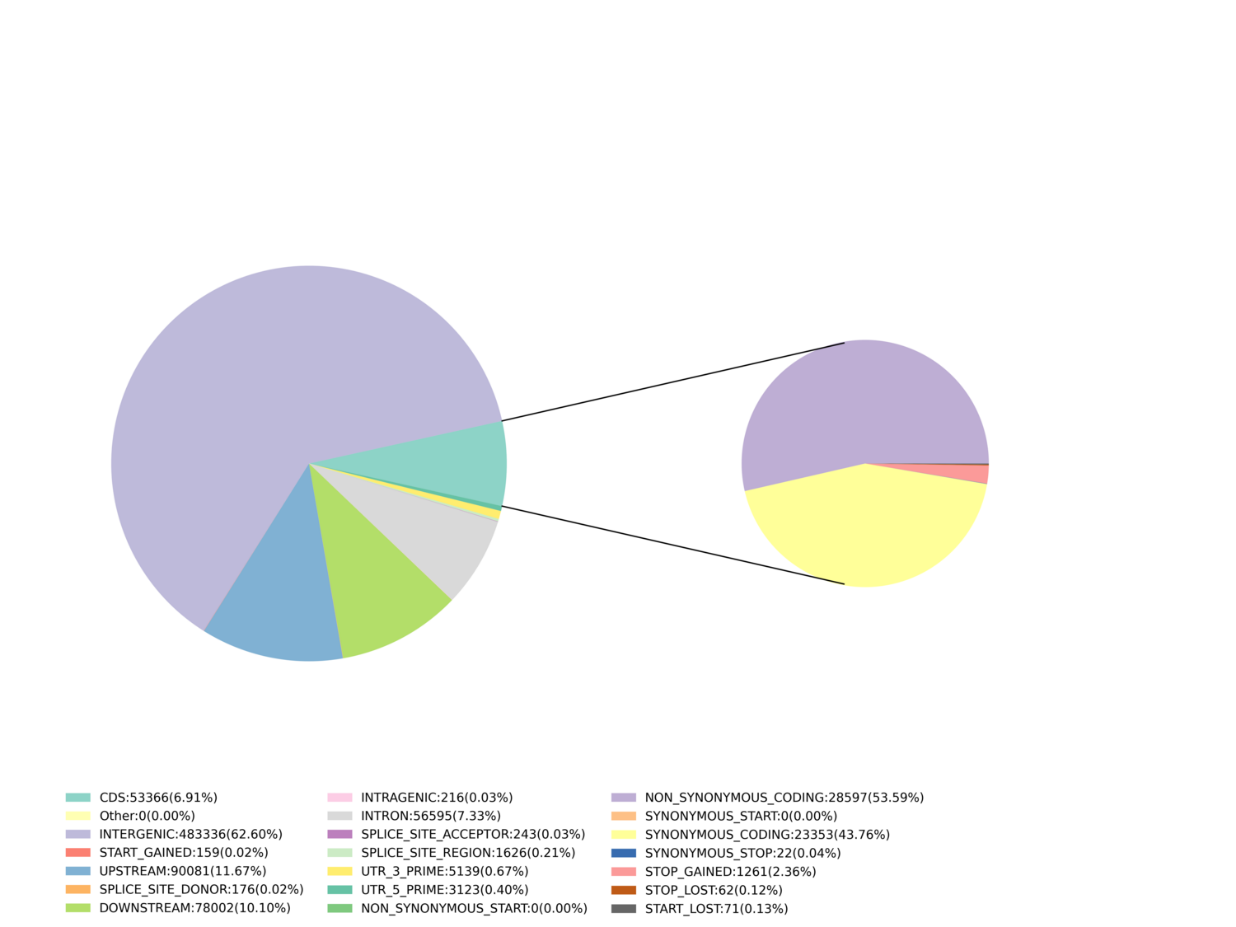
- Venalicium Inventionis: SNP, Indel, SNV, CV vocant et annotation
Distribution of Tags in chromosomes:
Distribution of Snps in chromosomes:
| Annus | Journal | IF | Titulus | Applications |
| MMXXII | Natura Communications | 17,694 | Genomic ex Giga-Chromosomes et Giga-genome of arbore AGLAY Paeonia Ostii | Slaf, Gwas |
| MMXV | Novum phytologist | 7,433 | Domesticas vestigia ancoris genomic regiones agronomic momenti in soybeans | Slaf, Gwas |
| MMXXII | Journal de Advanced Research | 12,822 | Genome-wide artificialis intravertam Gossypium barbadense in G. Hirsutum revelare superior loci simultaneous emendationem bombacio alimentorum fibra qualis et cede LAQUEUS | Slaf-Evolutionary Genetics |
| MMXIX | MOLECULA | 10.81 | Population genomic Analysis et de Novo Conventus revelare originem de Rapidshare Rice ut evolutionary ludum | Slaf-Evolutionary Genetics |
| MMXIX | Natura Genetics | 31.616 | Genome Sequentia et geneticae diversitatem communi carp, Cyprinus CARPIO | Slaf-Map |
| MMXIV | Natura Genetics | 25,455 | Genome coluerunt peanut praebet prudentiam in legumen Karyotypes, Polyploid Evolution et seges domesticae. | Slaf-Map |
| MMXXII | Plant Biotechnology Journal | 9.803 | Identificatio ST1 reveals a lectio involving Hitchhiking semen Morphology et oleum contentus durante soybean domesticae | Slaf-titulum progressionem |
| MMXXII | Journal of Acta Sciences | 6,208 | Identificatio et DNA venalicium progressionem ad triticum-Leymus molles 2ns (2d) Disomic parem substitutio chromosoma | Slaf-titulum progressionem |
| Annus | Journal | IF | Titulus | Applications |
| MMXXIII | Fines in planta scientia | 6,735 | Qtl Mapping et Transcriptome Analysis De Sugar Content in fructum maturis Pyrus Pyrifolia | GENICULUS |
| MMXXII | Plant Biotechnology Journal | 8,154 | Lepidium sativum de ST1 reveals a lectio involving Hitchhiking of semen Europologiam et oleum contentus durante soybean domesticae
| SNP vocatio |
| MMXXII | Fines in planta scientia | 6,623 | Genome-wide consociatio mapping of Hullless vix phenotypes in siccitatis elit.
| Gwas |