Totum genus resequensing

Genomics Cras Sars-Cov-II uncovers An NSP1 deletionem variante quod modulatas typus i et interferon responsio
Nanopore | Illumina | Totum genus Resequentia | Metagenomics | RNA, seq | Sanger
Technical Support in sample sequencing biomarkies technologies provisum provisum.
Elucidat
1.Sarsar-Cov-II Genome sequencing et phylognetic Analysis identify XXXV recurrentes mutationes inter XXXI Snps et IV.
2.Association cum CXVII orci phenotypes reveals in potentia
magna mutationes.
Δ500-532 in NSP1 coding regionis correlates cum inferioribus virales
3.Load et Serum ifn-β.
4.viral isolates cum Δ500-532 mutationem inducere inferior ifn-i
RESPONSIO in infectis cellulis.
Experimentalem consilio
Gestae
I. Covi-XIX epidemiological et genomic custodia
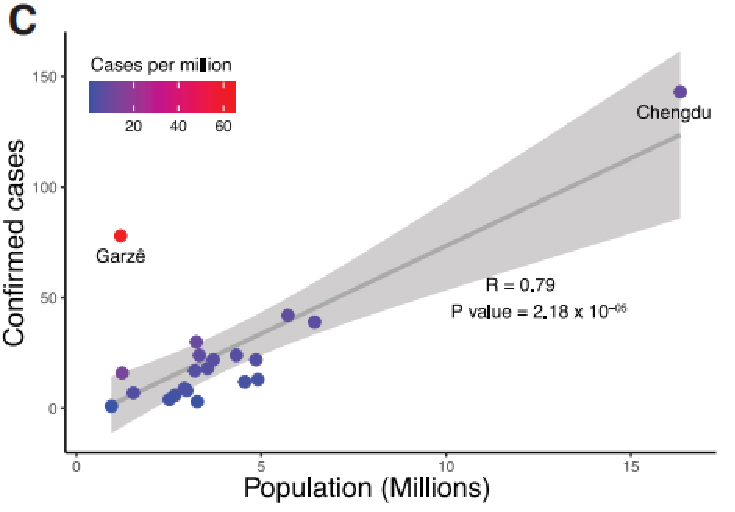
C. Data colligitur Sichuan provinciae, Sinis trans motum tempus a Jan. 22, MMXX ad Febr 20th, MMXX. A totalis DXXXVIII in Sichuan, 28.8% de qua ex provincia Capital. Confirmavit casibus in Sichuan auctus exponentially, Peaking on Jan. 30th. Item, data valet quod socialis perturbat potest esse a key factor in prohibendo virus propagationem.
Figura I. Epidemiological Study of Covi-XIX in Sichuan Province, Sina
II. Sars, Cov-II Genome constructione et variants idem
Cum multiplex PCR amplificatio sequitur Nanopore sequencing, a summa CCCX near- vel partialis, integrum genomes ex CCXLVIII aegris sunt generatae cum proxime. LXXX% of genomes operuit per X legit (medium profundum: 0.39 m legit per sample).
Figura II. Frequency cuiusque variants in Sichuan cohors
A summa CIV Snps et XVIII indels sunt identified a Sars, Cov-II genomes, in quo XXXI Snps et IV indels sunt identified quod recurrentes geneticae variants. Per comparet cum CLXIX exempla ex Wuhan et cum 81.391 summus qualitas Publiam Genome sequentia in Gisaid, XXIX de XXXV variants inventum presented in aliis continents. Notabiliter quatuor variants possidet δ500-532, Acc18108at, Δ729-737 et T13243C, tantum invenitur in Sichuan et Wuhan et absentes in Gisaid data, indicando quod variants erant valde probantes quod variants erant valde probare quod variantibus fortis, quae occursum Travel records de aegris.
Evolutionary analysis cum maximum likelihood (ml) modum et Bayesian Molecular horologium appropinquat et processionaliter in LXXXVIII Novum Virus Sfrom Sichuan et CCL curated genomes ab aliis regionibus. Genomes cum Δ500-532 (Deletions in NSP1 coding Regio) inventi sunt divisa sparsely in phylogenetic ligno. Haplotype Analysis in NP VI variants identified V ex illis ex multa urbes. Haec results suggesserant ut Δ500-532 occurrit in plures urbes et importari plures tempora ex Wuhan.
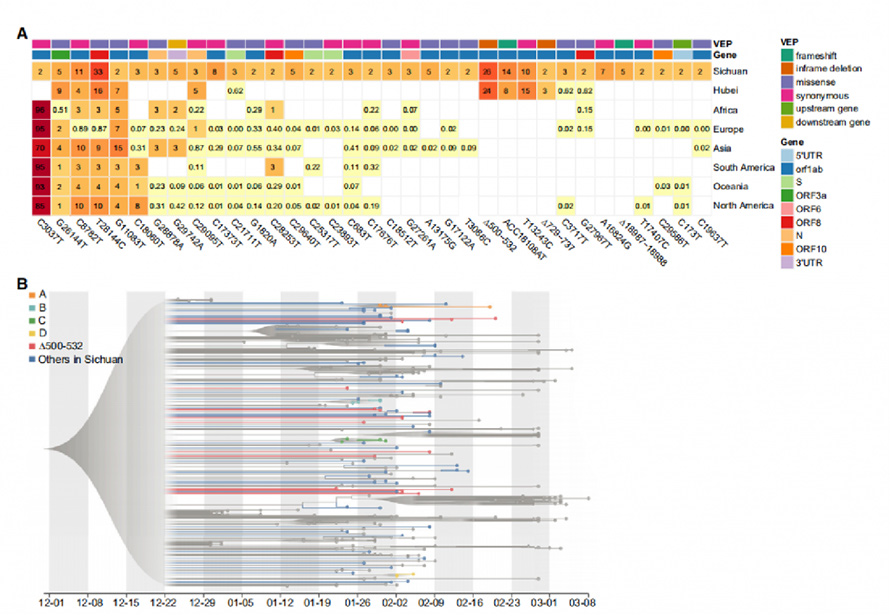
Figura II. Recurrent geneticae variants et phylogenetic Analysis in Sars, Cov- II genomes
III. Association of Recurrent geneticae variants cum orci effectus
CXVII orci Phenotypes cum COENTUM-XIX severitatem, ubi XIX severitatem actis phenotypes fuerunt classificatis in gravibus et non-gravi. Relatione inter haec lineamenta et XXXV recurrentes geneticae variants viualized in bi-botrum portet Heatmap. A GSEA, sicut ordo locupletantes analysis ostensum est quod Δ500-532 est negative connectuntur cum ESR, Serum iffn-β Andcd3 + CD8 + T cellula comitibus in sanguine. Praeterea, QPCR probat ostensum est quod aegris infici cum virus rapientur Δ500-532 habebat summum CT valore, id est lowest viral onus.
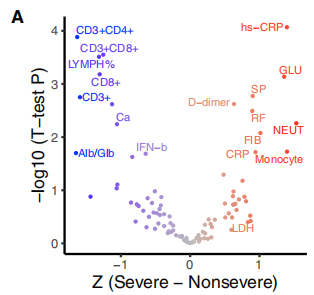
Figura III. Consociationes de XXXV recurrent geneticae variants cum orci pholidae
IV. Validation De Viral Mutationem consociata orci phenotypes
Ut intellegere afficit Δ500-532 in NSP1 munera, hek239t cellulis sunt transiit plasmids expressing plenus-longitudo, cum nsp1 et mutant formae deletionum. Transcriptome profiles cuiusque tractatus hek239t cellulis sunt processionaliter ad PCA analysis, showing quod deletionem mutants clusters respective propius et significantly diversis a wt nsp1. Et genes, quae significantly upregulated in mutants erant maxime locupletati sunt in "peptide biosynthetic / metabolicae processus", "Ribonucleoprotein complexu biogenesis", etc. et duo deletionum ostendit distincta expressa exemplar a Wt.
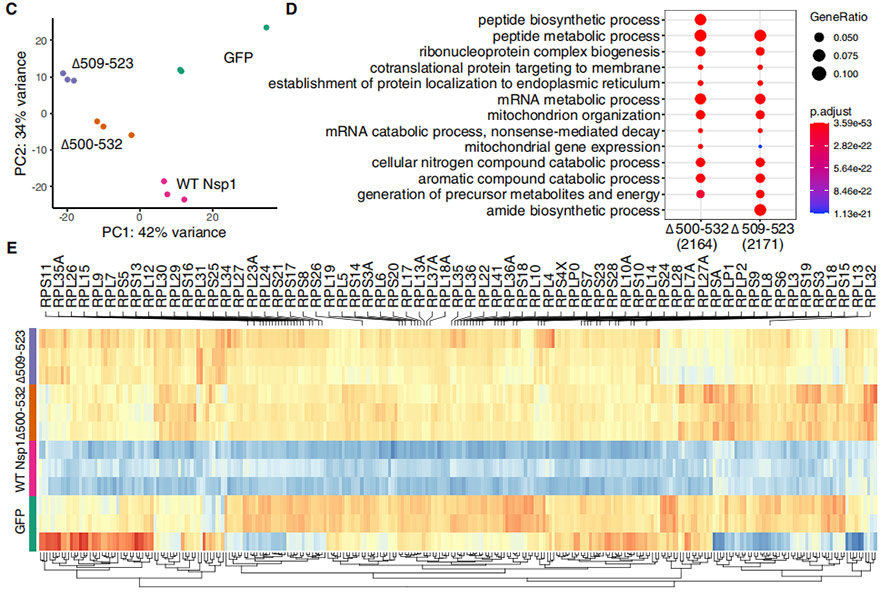
Figura IV. Transcriptome in HeK239T cellulis a WT NPS1 et cum deletionum
Afficit Deletions on IFN, I responsionem erat etiam probata in overexpressed studio. Omnes probata Deletions ostensum est ad redigendum iffn-I repsopse in transcenderunt hek239t et A549 cellulis ut tum transcriptome gradu et dapibus gradu. Interestingly, the significantly down-regulated genes in deletions were enriched in “defense response to virus”, “viral genome replication”, “regulation of transcription by RNA polymerase II” and “response to type I interferon”.
Figura V. Down DISCIPULUS Interferon Signaling Meatus in δ500-532 Mutant
In hoc studio, impulsum horum deletionum super virus ulteriores confirmati a viral infectio studiis. Virus per quaedam mutants erant separatim ex orci exemplaria et infici ad Calu-III cellulis. Detailed Results in viral infectio studio potest legere in charta.
doi:10.1016 / j.chom.201.01015
Referatio
Lin J, Tang C, Wei H, et al. Genomic Cras Sars-Cov-II uncovers an NSP1 deletionem variant quod modulat typus i interferon responsum [J]. Cell Host & Microbe, MMXXI.
Nuntium et ornare ipsum Aims ad sharing ad tardus felix casibus cum biomarker technologiae, caperent nova scientific res gestae tum prominentibus techniques applicari per studium.
Post tempus: Ian-06-2022


