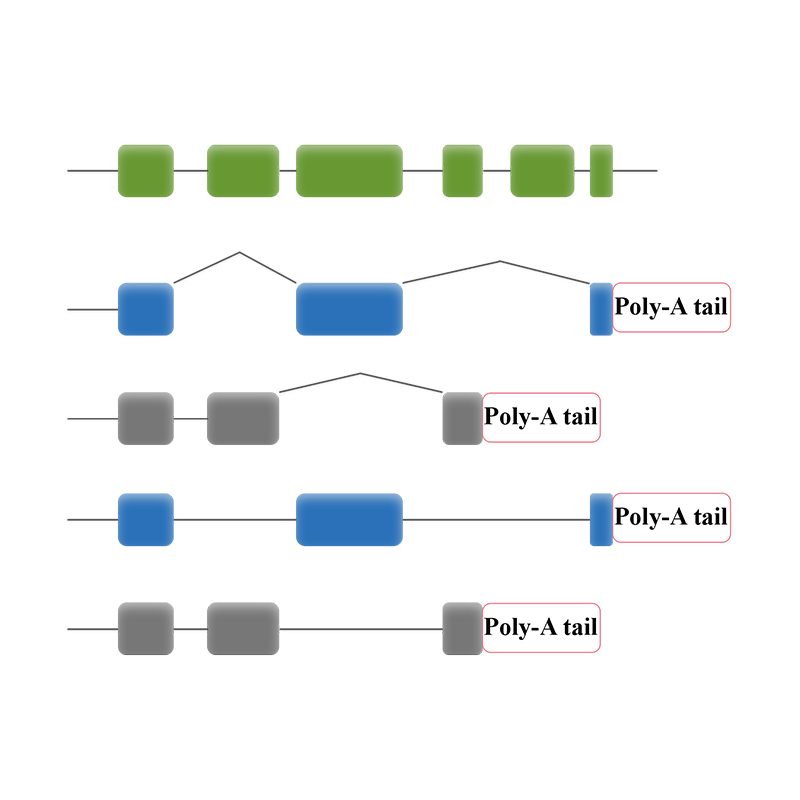
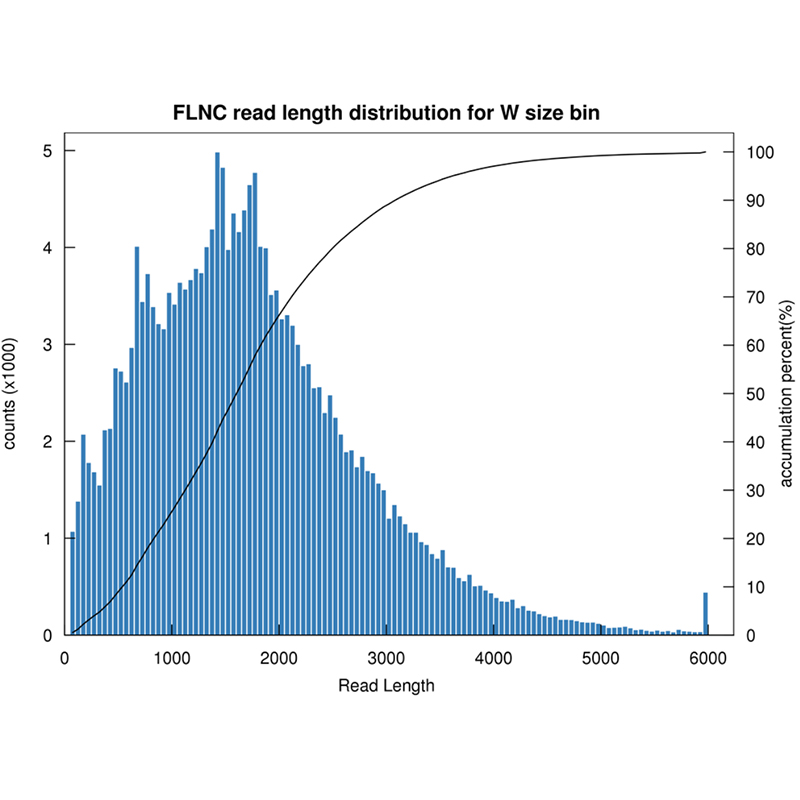
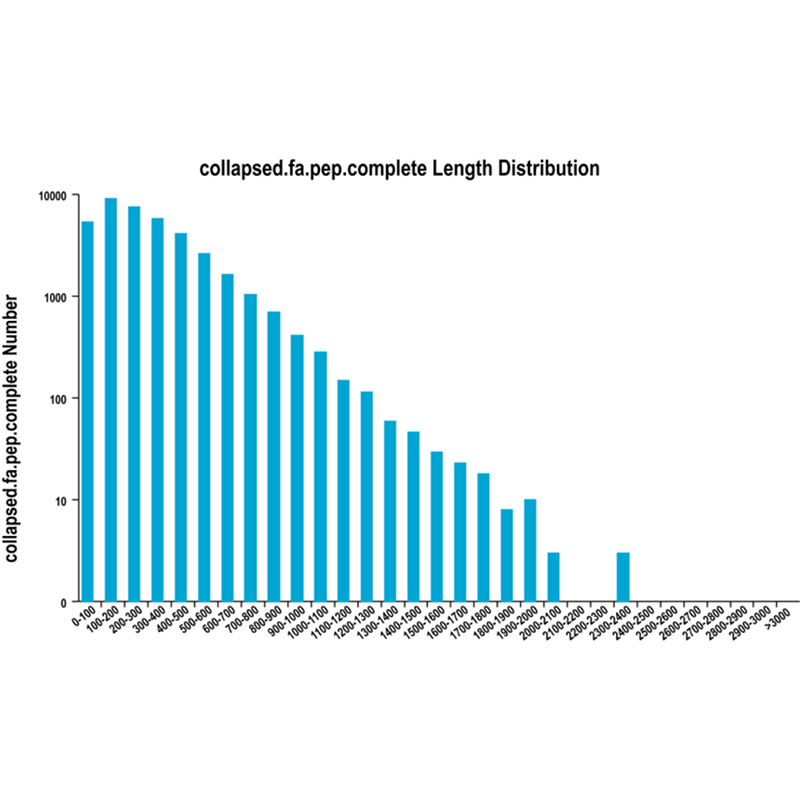
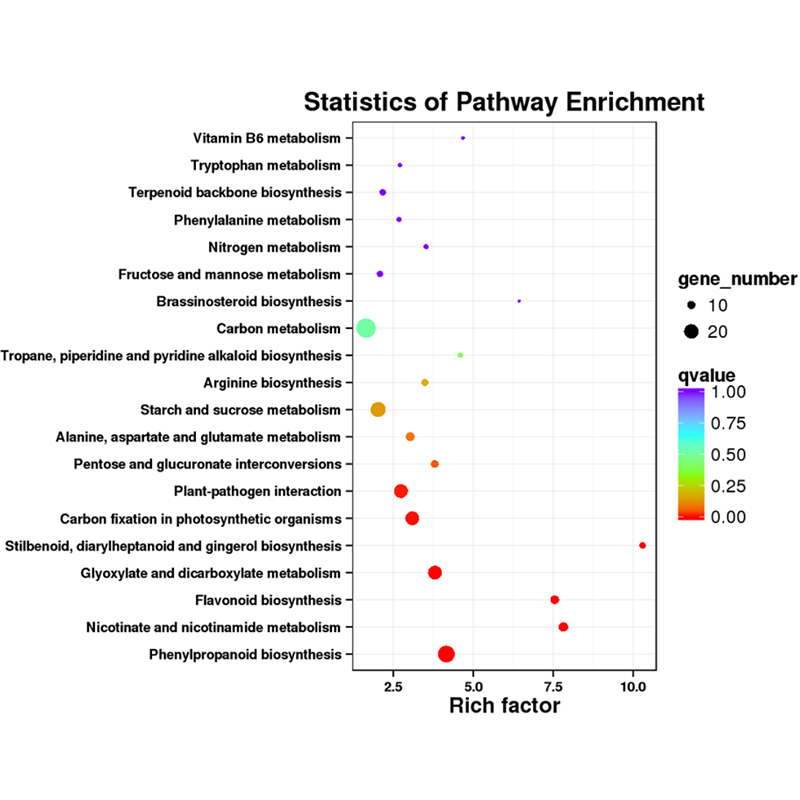
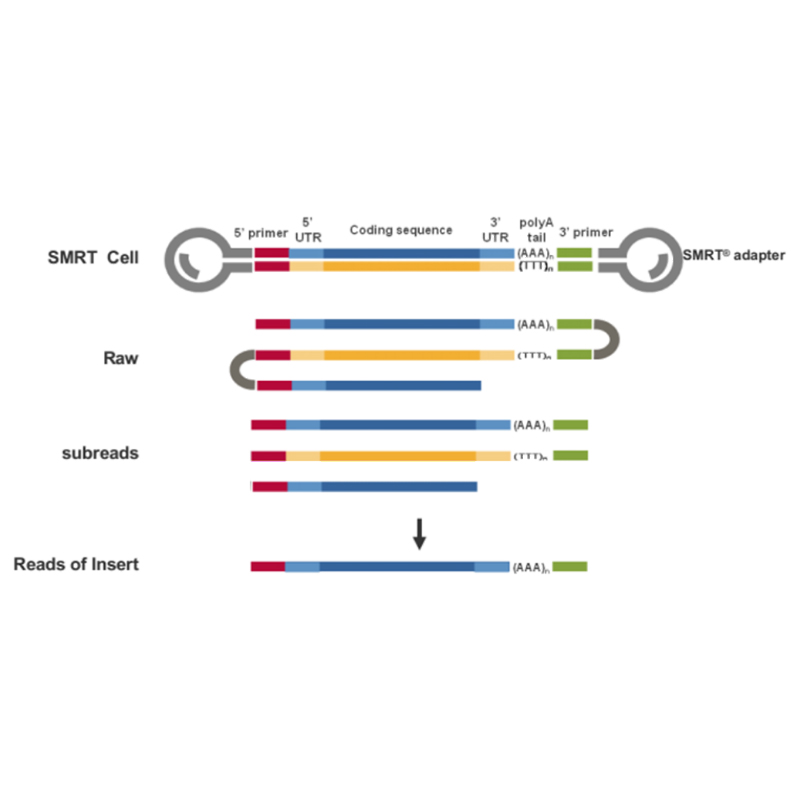
Plena longitudinem variant -PacBio . sequencing
Features
cDNA synthesis e poly-A mRNA sequitur praeparatio bibliothecae
● Sequencing in CCS modus, generandi HiFi legit
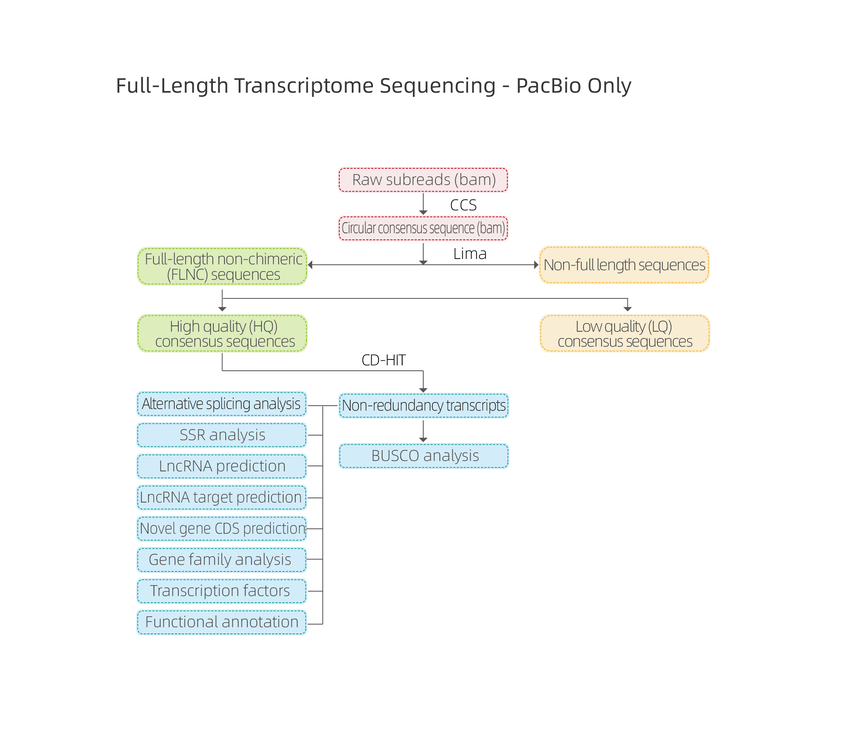
Sequencing plenae longitudinis transcripts
Analysis genome relationem non requirit; tamen adhiberi potest
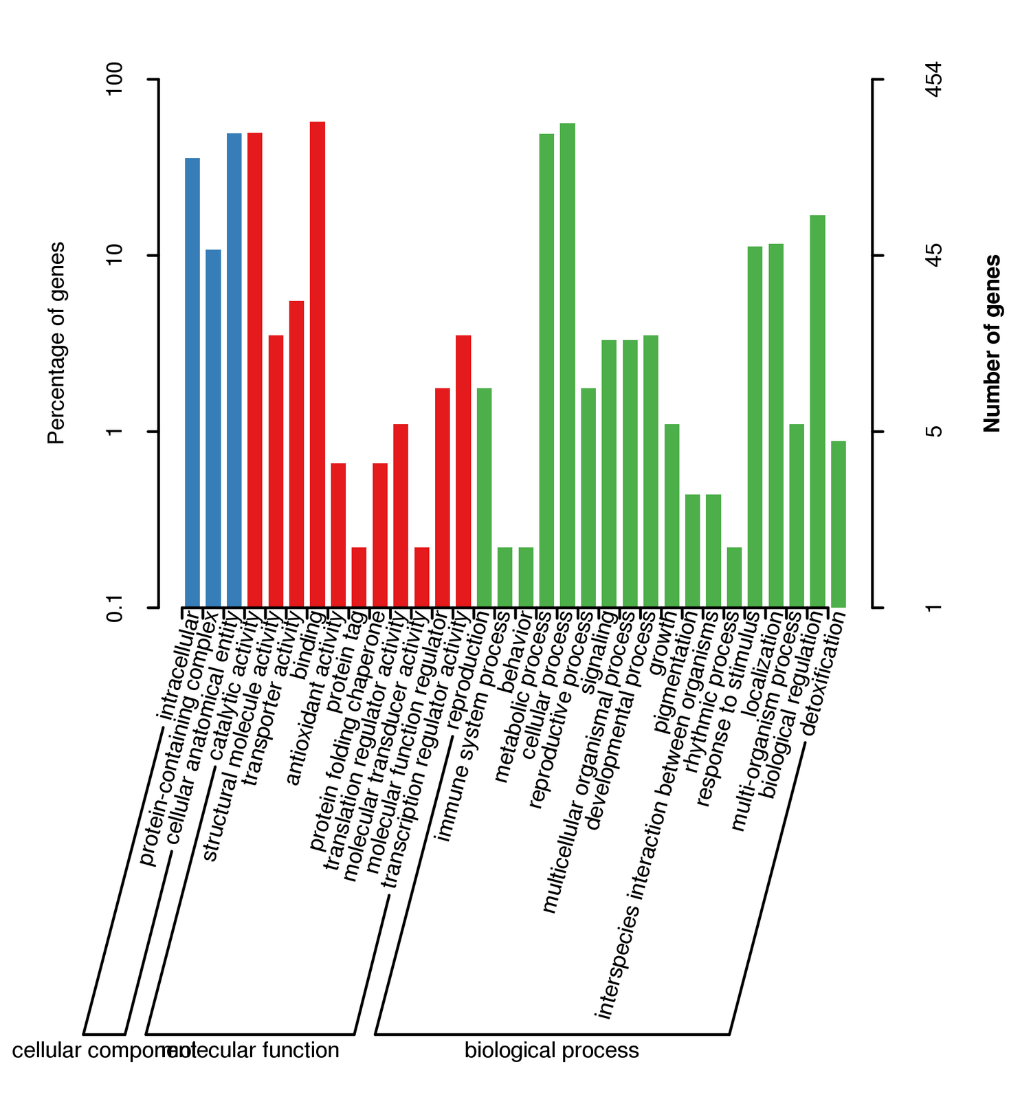
● Analysis bioinformatica dat analysin transcripts isoformes lncRNA, fusions gene, adenylation poly-adenylation, structura gene
Servitium Commoda
●Princeps Accuracy: HiFi accurate legit >99.9% (Q30), comparandum cum NGS
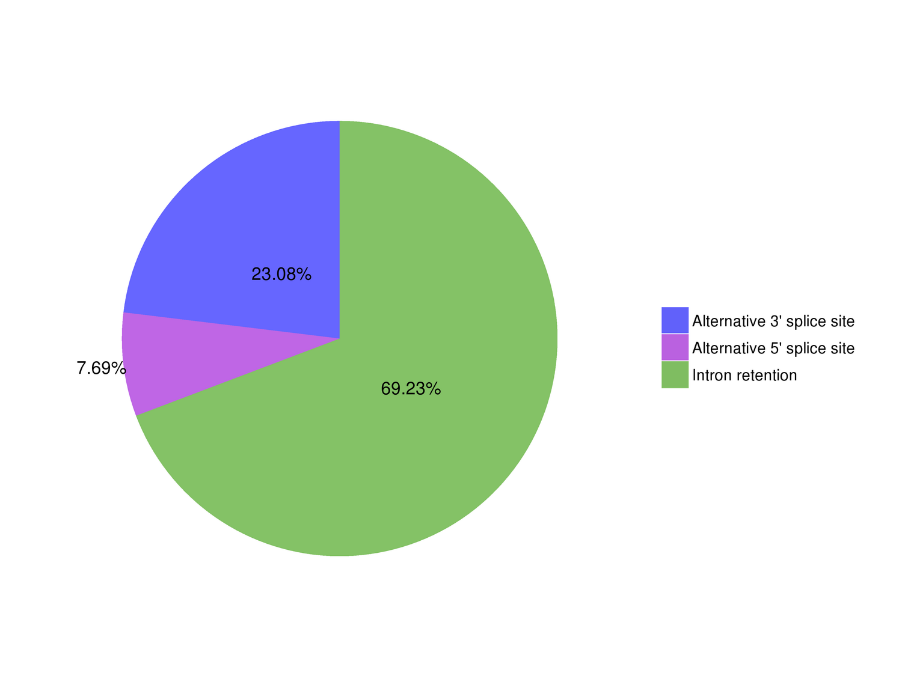
Alternative Splicing Analysis: sequencia totius transcriptorum dat isoform identificatio et characterisation
●Expertise: cum indagatio peractae super 1100 PacBio incepta plena longitudinis transcripto et dispensando super 2300 exempla, nostra turma experientiae opes ad omne propositum affert.
●Post-Sales Support: munus nostrum ultra project complementum cum 3-mensibus post venditionis tempus extenditur. Hoc tempore consilium praebemus subsequens, adiumentum fermentum, et sessiones Q&A address quaelibet interrogationes ad eventus relatas.
Sample Requisita et Delivery
| Library | Sequencing strategy | Data commendatae | Qualitas Imperium |
| PolyA ditavit variant CCS bibliothecae | PacBio Sequel II PacBio Revio | 20/40 Gb 5/10 M CCS | Q30≥85% |
Sample Requisita:
Nucleotides:
● Plantae:
Radix, Caulis vel Petal: 450 mg
Folium vel Semen: 300 mg
Fructus: 1.2 g
Animal:
Cor vel Intestinum: 300 mg
Viscera vel cerebrum: 240 mg
Musculus: 450 mg
Ossa, capillus vel cutis: 1g
Arthropoda:
Insecta: 6g
Crustacea: 300 mg
totum sanguinem: 1 tube
Cellulae: 106 cellulae
| Conc. | Moles (μg) | Puritas | Integritas |
| ≥ 100 | ≥ 1.0 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Nullam dapibus vel dapibus vel DNA contagione gel ostensum est. | Pro plantis: RIN≥7.5; Pro animalibus: RIN≥8.0; 5.0≥ 28S/18S≥1.0; limitata vel non collocantur elevatione |
Sample Delivery commendatur
Continens: 2 ml tubum centrifugium (foil plumbi non commendatur)
Sample labeling: Group+replicata eg A1, A2, A3; B1, B2, B3.
Shipment:
1. Siccum-glacies: Exemplaria in saccis referta necesse est et in glacie siccitate sepulta.
2. RNAstable tube: RNA specimina in RNA stabilizationis tubo exsiccata possunt (eg RNAstable®) et in cella temperatura exsiccata sunt.
Service Opus O

Experimentum design

Sample traditio

RNA extractionem

Library construction

Sequencing

Analysis Data

Post-venditionis officia
Sequenti analysi includit:
Rudis notitia qualis imperium
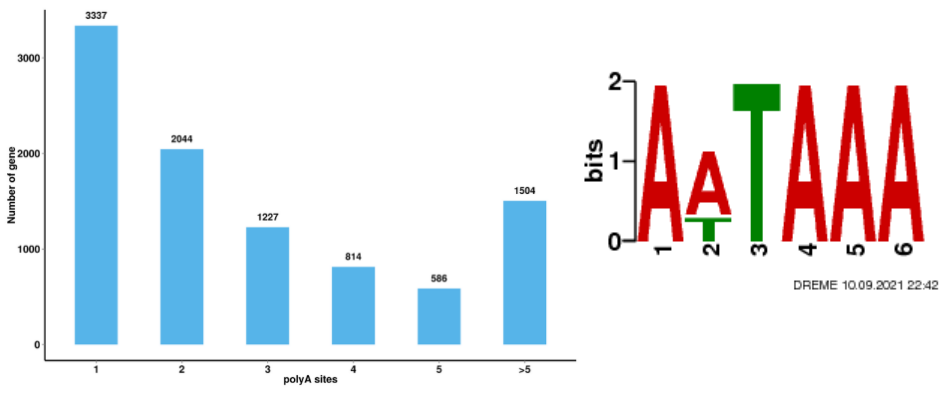
Alternative Polyadenylation Analysis (APA)
Fusion transcript analysis
Alternative Splicing Analysis
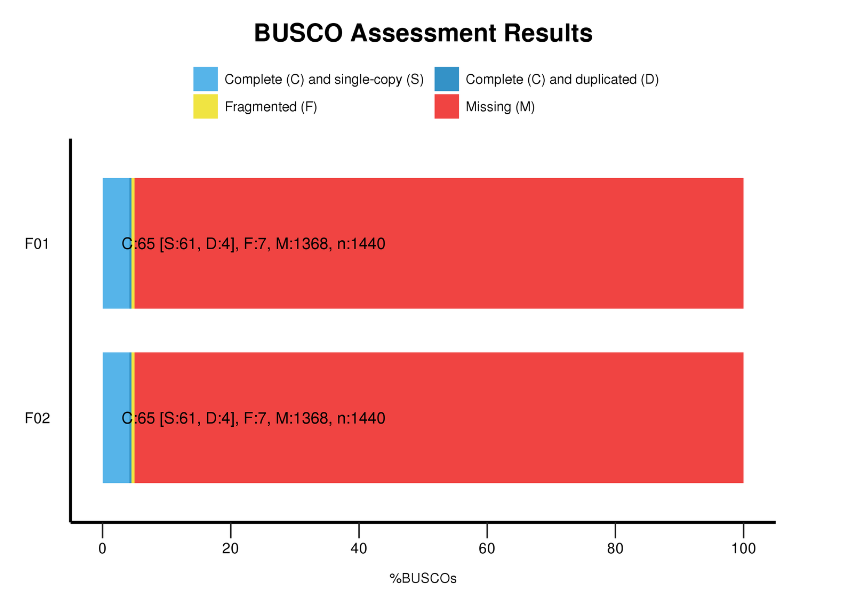
Benchmarking Universal Single Exemplar Orthologorum (BUSCO) analysis
● Analysis Novella transcript: praesagium sequentiarum coding (CDS) et annotationem functionis
lncRNA analysis: vaticinium lncRNA et scuta
MicroSatelite Lepidium sativum (SSR)
BUSCO analysis
Alternative Splicing Analysis
Alternative Polyadenylation Analysis (APA)
Functional annotation of novel transcripts
Explorarent progressiones faciliores per BMKGene's Nanopore plenam-longitudinem mRNA sequencing officia in hac publicatione featured.
Ma, Y. et al. (2023) 'Comparative analysis of PacBio and ONT RNA sequencing methodos Nemopilema Nomurai veneni identificatio', Genomics, 115(6), p. 110709. doi: 10.1016/J.YGENO.2023.110709.
Chao, Q. et al. (2019) 'Motiones evolutionis transcriptome populi Populi, Plantarum Acta Biotechnologia, 17 (1), pp. 206-219. doi: 10.1111/PBI.12958.
Deng, H. et al. (2022) ' Mutationes dynamicae in Ascorbico Acidi contenti per progressionem fructuum et maturationem Actinidiae latifoliae (Ascorbate-Rich Fruit Crop) et Mechanismi associati Moleculares, Acta Scientiarum Molecularium Internationalium, 23(10), p. 5808. doi: 10.3390/IJMS23105808/S1.
Hua, X. et al. (2022) 'Praedictio efficax generum viarum biosyntheticae in polyphyllo bioactivo implicatae in polyphylla Parisiensi', Communicationis Biologiae 2022 5, 1, 5(1), pp. doi: 10.1038/s42003-022-03000-z.
Liu, M. et al. (2023) 'PacBio Iso-Seq deducta et Illumina RNA-Seq Analysis Tuta absoluta (Meyrick) Transcriptome et Cytochrome P450 Genes, Insects, 14 (4), p. 363. doi: 10.3390/INSECTS14040363/S1.
Wang, Lijun et al. (2019) 'Aspectio transcriptome complexionis utens PacBio analysi uni-moleculi realis-tempus cum Illumina RNA, ad meliorem intelligentiam acidi ricinoleici biosynthesis in Ricinus communis', BMC Genomics, 20 (1), pp. 1-17. doi: 10.1186/S12864-019-5832-9.