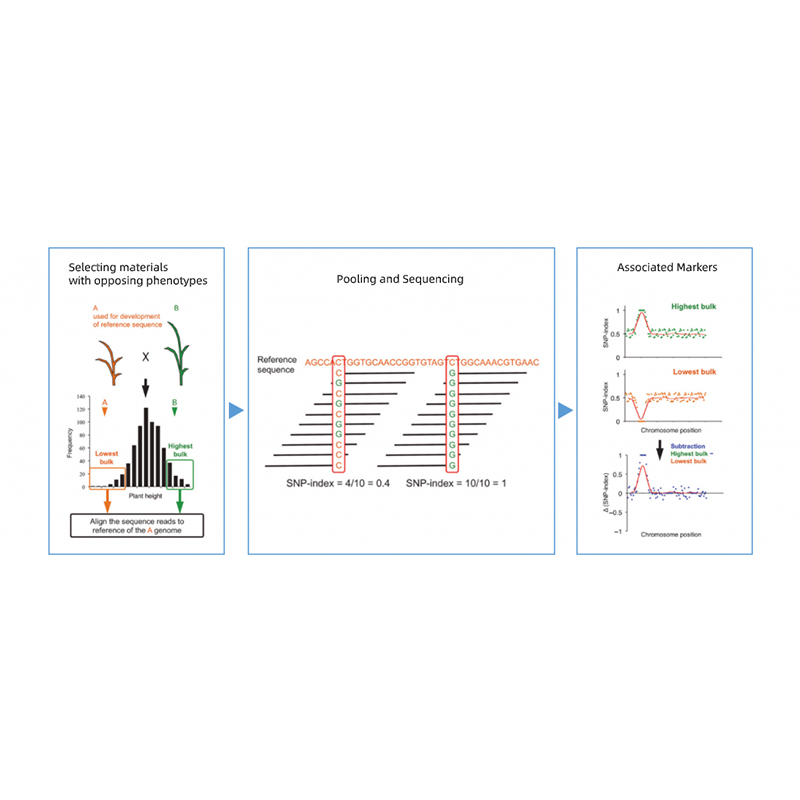
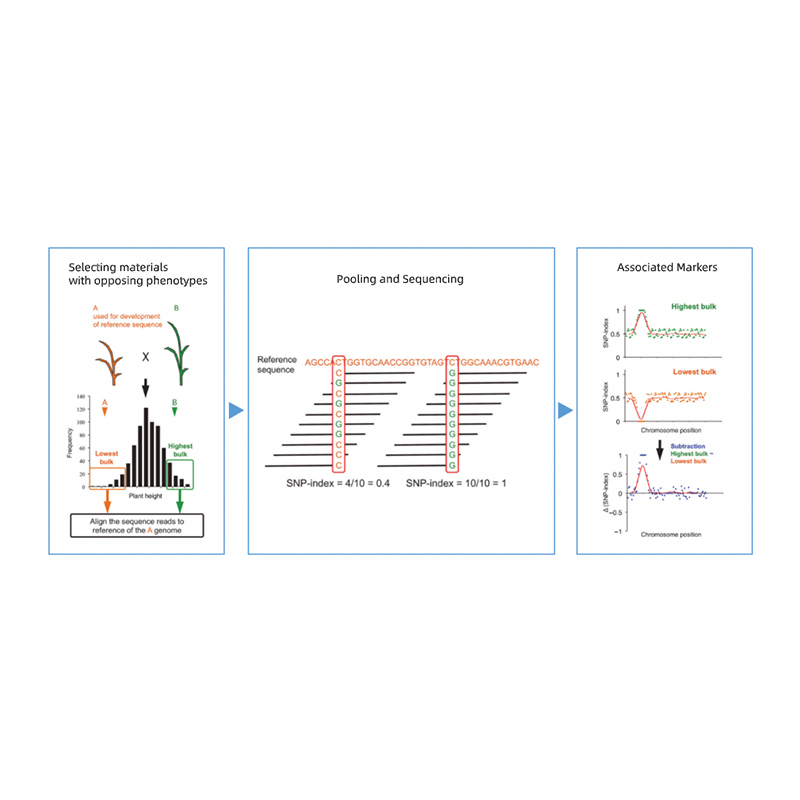
Bullked segregant analysis
Service commoda
Takagi et al., Plant Journal, MMXIII
● accurate localization: miscentes cum XXX + CC + CC + CC ad minimize background sonitus; non-synticas mutatants, secundum candidatum regione praedictionem.
● Aliquam Analysis: in-profundum candidatus gene munus annotation, inter NR, SwissProt, GO, KEGG, COG, KOG, etc.
● citius Turnaround tempus, celeri gene localization in XLV working days.
● extensive experientia: BMK has operam in millia of lineamenta localization, operientur diverse species sicut fruges, aquatilium products, silvam, flores, fruges, etc.
Service Specifications
Population:
Segregating progenies parentum contraria Phenotypes.
eg F2 Progeny, backcrossing (BC), Recombinant INBRED linea (RIL)
POOL
Qualitative Lineamenta: XXX ad L hominum (minimum XX) / mole
Nam quantitatis tratis: Top V% ad X% hominum cum aut extremum phenotypes in toto population (minimum XXX + XXX).
Commendatur sequencing profundum
Saltem 20x / parentis et 1x / fetus singulos (eg pro fetus miscentes piscinam XXX + XXX singula, sequencing profundum erit 30x per mole)
Bioinformatics analyzes
● totum genome resequentiora
● data processus
● SNP / Indel vocantem
● candidatus regionem protegendo
● Candidatus Gene munus annotation
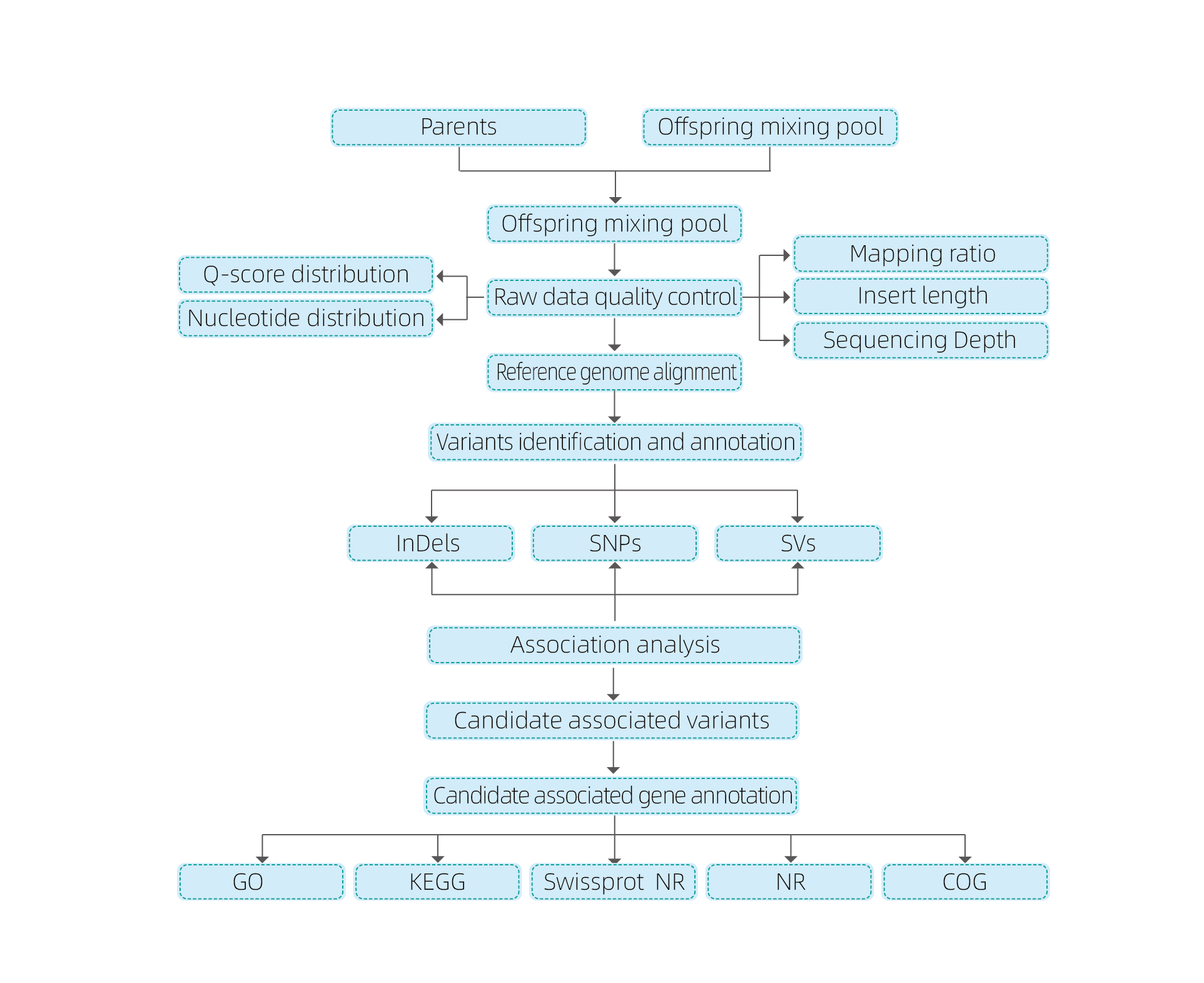
Sample requisitis et partus
Sample Requirements:
Nucleotides:
| GDNA Sample | TEXTUS Sample |
| Concentration: ≥3 / μl | Plantarum: 1-2 G |
| Moles: ≥2 μg (Volumn ≥15 μl) | Animalium 0.5-1 G |
| Puritas: OD260 / CCLXXX = 1.6-2.5 | Omnis sanguis: 1.5 ml |
Service opus influunt

Experimentum Design

Sample partum

RNA extraction

Constructio

Sequencing

Analysis

Post-Sale Services
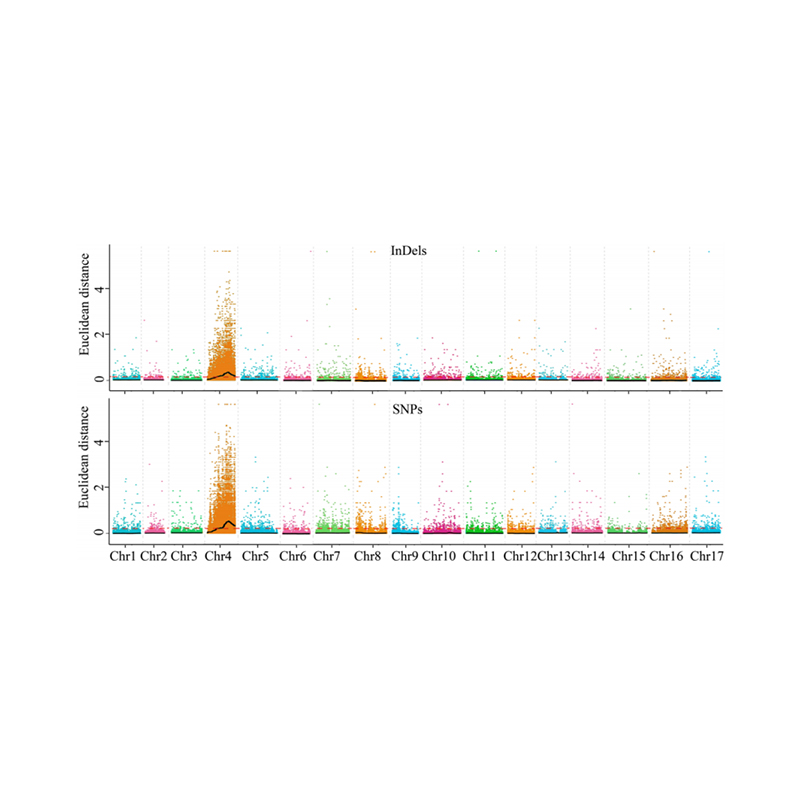
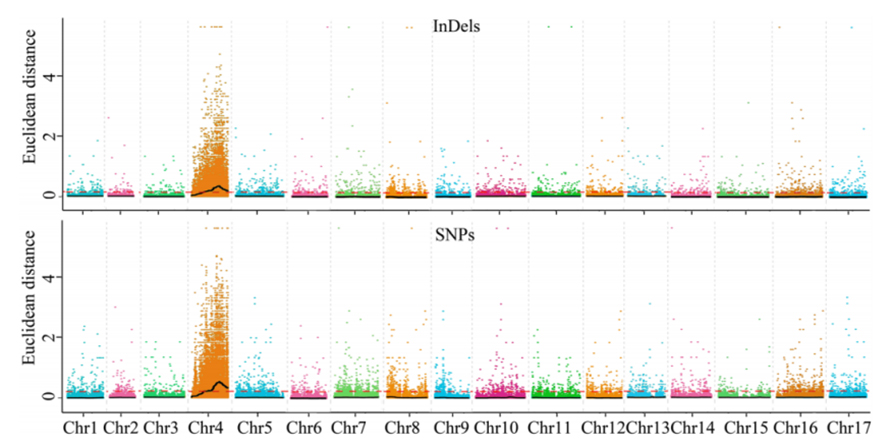
1.Association Analysis basi in Euclidean distantiam (ed) ad identify candidatum regionem. In his figure
X-axis: Chromosoma numero; Quisque dot repraesentat an Ed valorem de snp. Nigrum linea correspondent aptavit ed pretii. A superiore ed pretii indicat magis significant consociatio inter situm et phenotype. Rubrum Dash linea represents limen of significant consociatio.
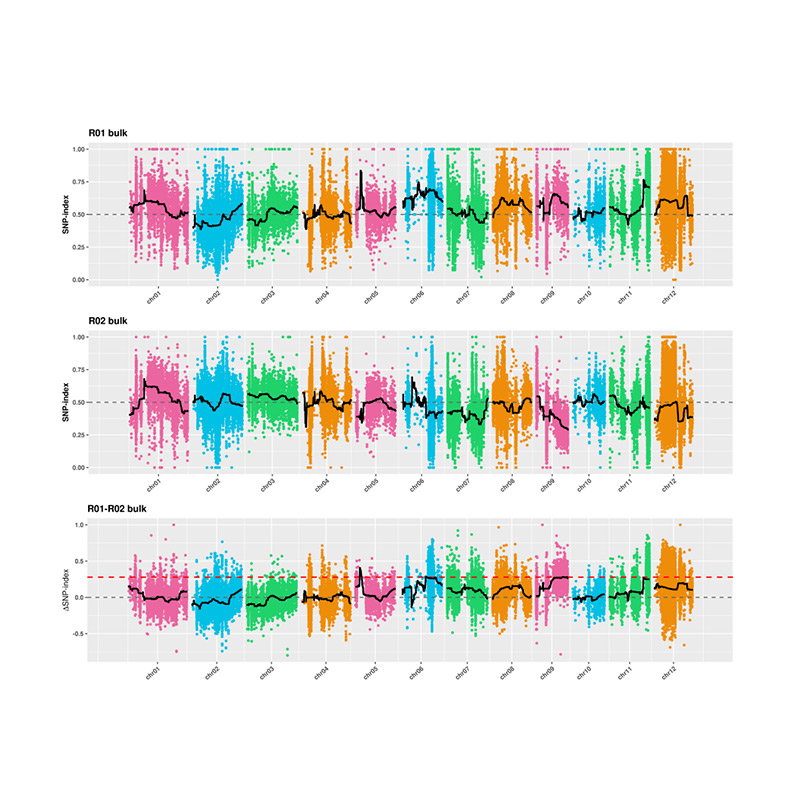
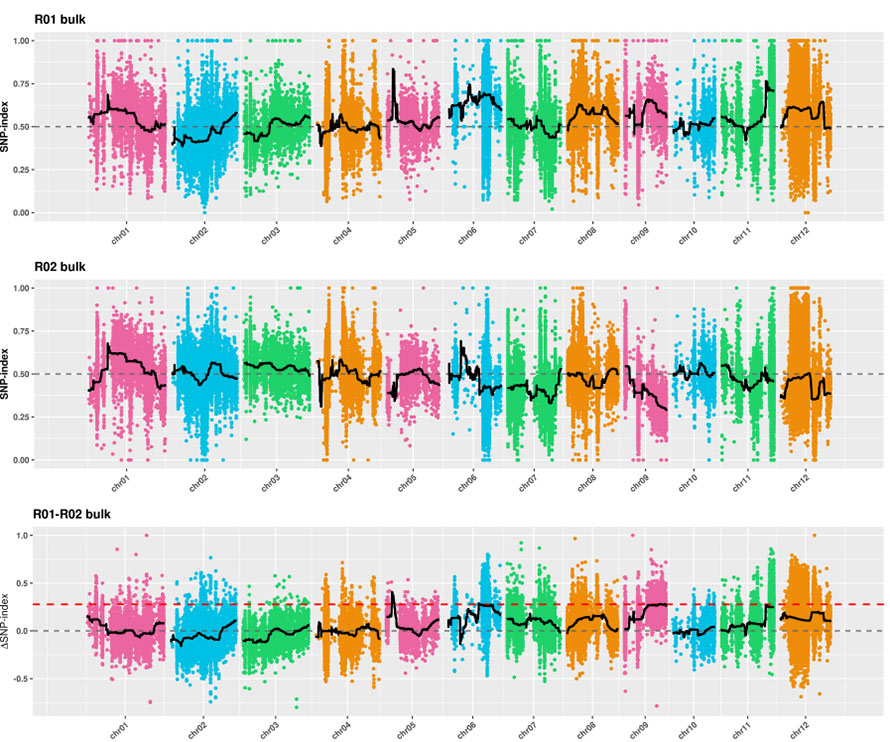
2.Association Analysis secundum Nemo SNP-Index
X-axis: Chromosoma numero; Quisque dot repraesentat snp-index pretii. Nigrum linea stat pro aptavit snp-index pretii. Quod maior est valor est, magis significant consociatio est.
BMK Case
Maior-effectus quantitativam lineamentum locus fnl7.1 encodes a nuper embryogenesis abundat dapibus consociata cum fructus collum longitudinem in cucumis
Published: Plant Biotechnology Journal, MMXX
Sequencing Strategy:
Parentes (Jin5-508, YN): tota genome resequentationem ad XXXIV × XX ×.
DNA Pools (L longa cervicis et L brevi cervice): Resequentcing ad LXI × et LII ×
Clavis results
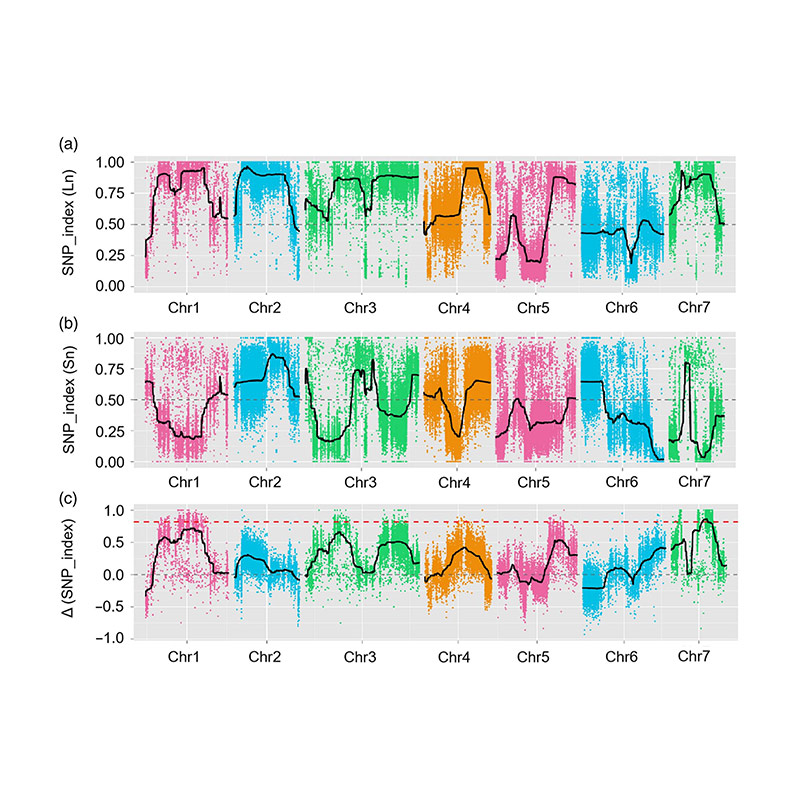
In hoc studio, Segregating population (F2 et F2: III), quod generatae transitus longa-collo cucumis linea jin5-508 et brevis-collo y. Duo DNA lacus constructum L longa-collo hominum et L extremam brevis collum hominum. Maior-effectus Qtl est identified in Chr27 by BSA Analysis et Traditional QTL Mapping. In candidatus regione adhuc preterea ad denique-mapping, gene expressio quantitatem et transgenic experimenta, quae revelata key gene in moderantum collum-longitudo, csfnl7.1. Praeterea, Polymorphism in CSFNL7.1 Promotor regionem inventa esse consociata cum correspondentes expressio. Praeterea phylogenetic analysis suggesserant quod fnl7.1 locus est valde verisimile ad originem ex India.
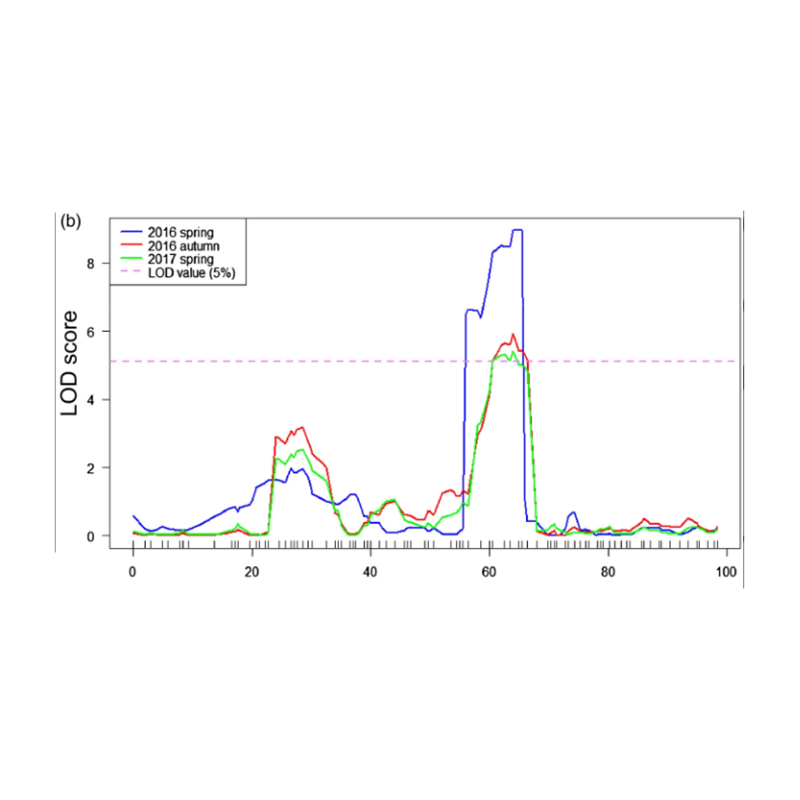
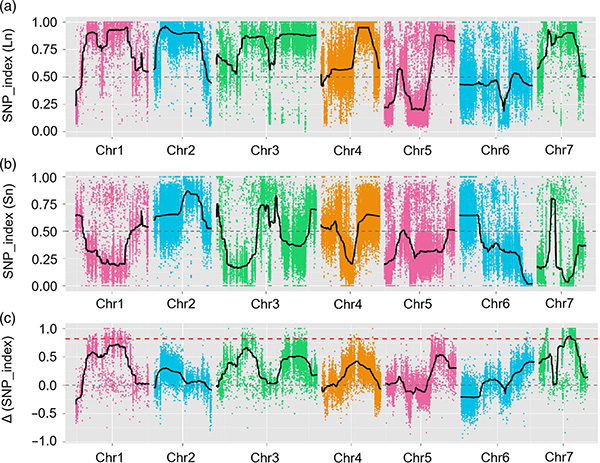 QTL-Mapping in BSA Analysis ad identify candidatum regionem consociata cum cucumis colli longitudinem | 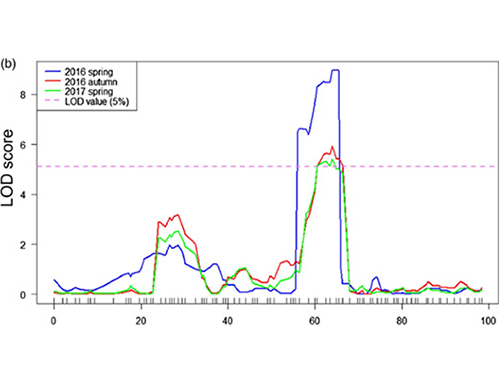 LOD Profiles cucumis colli-longitudo Qtl identified in Chr27 |
Xu X. et al. "Maior-effectus quantitativam lineamentum locus fnl7.1 encodes a nuper embryogenesis abundat dapibus consociata cum fructus collum longitudinem in cucumis." 18,7 Plant Journal (MMXX).