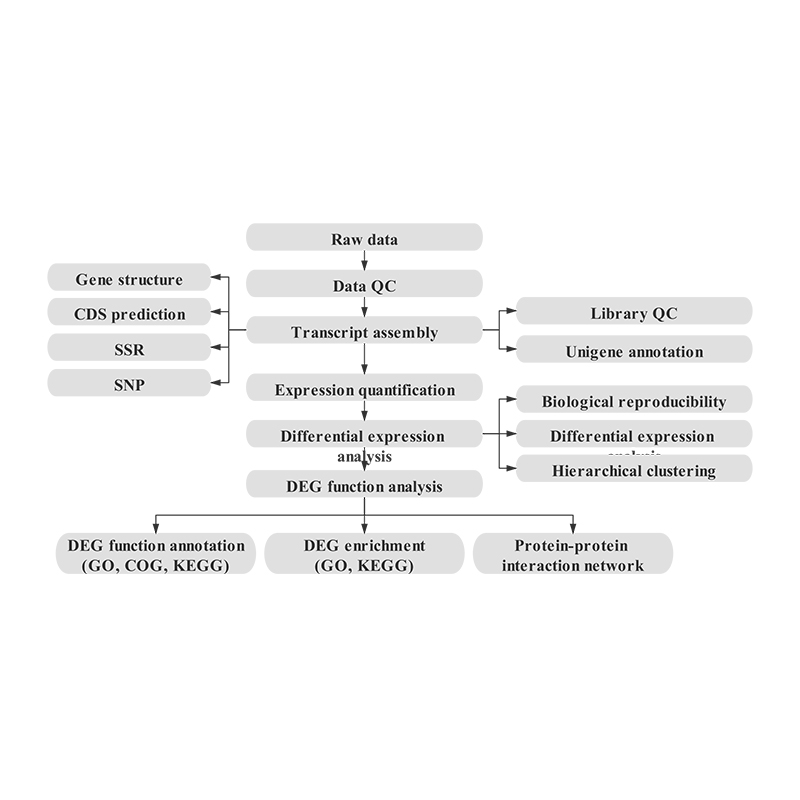
Usoro mRNA na-abụghị akwụkwọ ntụaka-NGS
Atụmatụ
● Ijide poly mRNA tupu nkwadebe ọbá akwụkwọ
● N'adabereghị na genome ọ bụla: dabere na mgbakọ de novo nke transcripts, na-emepụta ndepụta unigenes nke ejiri ọtụtụ ọdụ data kọwaa (NR, Swiss-Prot, COG, KOG, eggNOG, Pfam, GO, KEGG)
● Nyocha bioinformatic zuru oke nke okwu mkpụrụ ndụ ihe nketa na nhazi transcript
Uru ọrụ
●Ọkachamara sara mbara: na ndekọ egwu nke nhazi ihe karịrị 600,000 samples na BMKGENE, na-agbasa ụdị ụdị dị iche iche dị ka omenala cell, anụ ahụ, na mmiri ara, ndị otu anyị na-eweta ahụmahụ bara ụba na ọrụ ọ bụla. Anyị emechiela ihe karịrị 100,000 mRNA-Seq ọrụ na ngalaba nyocha dị iche iche.
●Njikwa ogo siri ike: anyị na-emejuputa isi ihe nchịkwa n'ofe niile, site na nlele na nkwadebe ọbá akwụkwọ ruo usoro na bioinformatics. Nleba anya nke ọma a na-eme ka o doo anya na nnyefe nke nsonaazụ dị elu mgbe niile.
● Nkọwa zuru oke: anyị na-eji ọtụtụ ọdụ data na-arụ ọrụ ịkọwapụta Genes dị iche iche (DEGs) na-arụ ọrụ ma na-eme nyocha nke ịba ụba kwekọrọ, na-enye nghọta na usoro cellular na molecular na-akpata nzaghachi transcriptome.
●Nkwado maka ire ere: nkwa anyị ga-agafe karịa mmecha ọrụ yana oge ọrụ ọnwa 3 gachara. N'ime oge a, anyị na-enye nlebanya ọrụ, enyemaka nchọpụta nsogbu, yana oge ajụjụ & A iji zaa ajụjụ ọ bụla metụtara nsonaazụ ya.
Ihe nlele chọrọ na nnyefe
| Ụlọ akwụkwọ | Usoro usoro | akwadoro data | Njikwa ogo |
| Poly A bara ụba | Ilumina PE150 DNBSEQ-T7 | 6-10 Gb | Q30≥85% |
Ihe atụ chọrọ:
Nucleotides:
| Conc.(ng/μl) | Ọnụ ego (μg) | Ịdị ọcha | Iguzosi ike n'ezi ihe |
| ≥ 10 | ≥ 0.2 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Enwere oke ma ọ bụ enweghị protein ma ọ bụ mmetọ DNA nke egosiri na gel. | Maka osisi: RIN≥4.0; Maka anụmanụ: RIN≥4.5; 5.0≥28S/18S≥1.0; nwere oke ma ọ bụ enweghị mgbago elu |
● Osisi:
Mgbọrọgwụ, azuokokoosisi ma ọ bụ petal: 450 mg
Akwụkwọ ma ọ bụ mkpụrụ: 300 mg
Mkpụrụ: 1.2 g
● Anụmanụ:
Obi ma ọ bụ eriri afọ: 300 mg
Viscera ma ọ bụ ụbụrụ: 240 mg
Akwara: 450 mg
Ọkpụkpụ, ntutu ma ọ bụ akpụkpọ: 1g
● Arthropods:
Ụmụ ahụhụ: 6g
Crustacea: 300 mg
● Ọbara dum: 1 tube
● Selụ: 106 mkpụrụ ndụ
Nnyefe nlele akwadoro
Akpa: 2 ml centrifuge tube (Tin foil adịghị akwadoro)
Ịkpọ aha nlele: Otu+ megharịa dịka A1, A2, A3; B1, B2, B3.
Mbupu:
1. Dry-ice: A ga-etinye ihe nlele n'ime akpa ma lie ya na ice-akọrọ.
2. tubes RNAstable: Enwere ike ịkpọsa ihe nlele RNA na tube stabilization RNA (dịka ọmụmaatụ RNAstable®) ma bufee ya n'ime ụlọ okpomọkụ.
Usoro ọrụ ọrụ

Nnwale imewe

Nbufe ihe atụ

mmịpụta RNA

Ịrụ ụlọ akwụkwọ

Usoro

Nyocha data

Mgbe-ire ọrụ
Bioinformatics
Mgbakọ transcriptome na nhọrọ unigene
Nkọwa nke Unigene
Ihe nlere anya na ntule nke ntugharị ndu
Mkpụrụ ndụ ihe nketa egosipụtara dị iche iche (DEGs)
Nkọwa arụmọrụ nke DEGs
Ọganihu arụ ọrụ nke DEGs
Nyochaa ọganihu ndị ọrụ nhazi usoro eukaryotic NGS mRNA BMKGene na-akwado site na nchịkọta mbipụta.
Shen, F. et al. (2020) 'Mgbakọ transcriptome De novo na mkpụrụ ndụ ihe gbasara mmekọahụ na gonads nke Amur catfish (Silurus asotus)', Genomics, 112(3), p. 2603–2614. doi: 10.1016/J.YGENO.2020.01.026.
Zhang, C. et al. (2016) 'Ntụle transcriptome nke sucrose metabolism n'oge bọlbụ ọzịza na mmepe na yabasị (Allium cepa L.)', Frontiers in Plant Science, 7 (September), p. 212763. doi: 10.3389/FPLS.2016.01425/BIBTEX.
Zhu, C. et al. (2017) 'De novo mgbakọ, njirimara na nkọwa maka transcriptome nke Sarcocheilichthys sinensis', PLoS ONE, 12 (2). doi: 10.1371/JOURNAL.PONE.0171966.
Zou, L. et al. (2021) 'De novo transcriptome analysis na-enye nghọta na nnabata nnu nke Podocarpus macrophyllus n'okpuru nrụgide salinity', BMC Plant Biology, 21(1), p. 1-17. doi: 10.1186/S12870-021-03274-1/ỌFỌNỤ/9.