Nyocha gbasara mkpokọta Genome
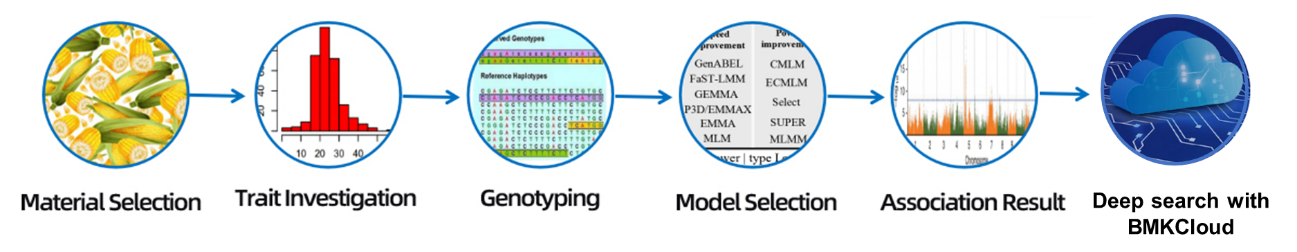
Usoro ọrụ
Uru ọrụ
●Ọkachamara sara mbara na ndekọ mbipụta: site na mkpokọta ahụmahụ na GWAS, BMKGene emechaala ọtụtụ narị ụdị ọrụ dị iche iche na nyocha GWAS nke ọnụ ọgụgụ mmadụ, nyere ndị nchọpụta aka ka ha bipụta ihe karịrị 100 akụkọ, ihe nchịkọta mmetụta ruru 500.
● Nyocha bioinformatics zuru oke: usoro ọrụ na-agụnye nyocha njikọ SNP-trait, na-ebuga usoro mkpụrụ ndụ ihe nketa na nkọwa ọrụ ha kwekọrọ.
●Ndị otu bioinformatics nwere nkà dị ukwuu na okirikiri nyocha dị mkpirikpi: nwere ahụmịhe dị ukwuu na nyocha genomics dị elu, ndị otu BMKGene na-enye nyocha zuru oke na oge ntụgharị ngwa ngwa.
●Nkwado mgbe ọrịre ahịa:Nkwenye anyị ga-agafe karịa mmecha ọrụ yana oge ọrụ ọnwa 3 gachara. N'ime oge a, anyị na-enye nlebanya ọrụ, enyemaka nchọpụta nsogbu, yana oge ajụjụ & A iji zaa ajụjụ ọ bụla metụtara nsonaazụ ya.
Nkọwapụta ọrụ na ihe achọrọ
| Ụdị usoro | Ọnụ ọgụgụ ndị akwadoro | Usoro usoro | Nucleotide chọrọ |
| Usoro genome niile | 200 sample | 10x | Ntinye uche: ≥ 1ng/µL Ngụkọta ego≥ 30ng Oke ma ọ bụ enweghị mmebi ma ọ bụ mmetọ |
| Iberibe Amplified Locus (SLAF) akọwapụtara nke ọma | Omimi mkpado: 10x Ọnụọgụ mkpado: <400 Mb: WGS ka akwadoro <1Gb: mkpado 100k 1Gb > 2Gb: mkpado 300k Mkpado kacha 500k | Ntinye uche ≥ 5 ng/µL Mkpokọta ego ≥ 80 ng Nanodrop OD260/280=1.6-2.5 Agarose gel: enweghị ma ọ bụ oke mmebi ma ọ bụ mmetọ
|
Nhọrọ ihe



Ụdị dị iche iche, ụdị subspecies, landraces / genebanks / ezinụlọ agwakọtara / akụ anụ ọhịa
Ụdị dị iche iche, ụdị ala, ala
Ezinụlọ ọkara-sib/ezinaụlọ zuru oke/ihe anụ ọhịa
Usoro ọrụ ọrụ

Nnwale imewe

Nbufe ihe atụ

mmịpụta RNA

Ịrụ ụlọ akwụkwọ

Usoro

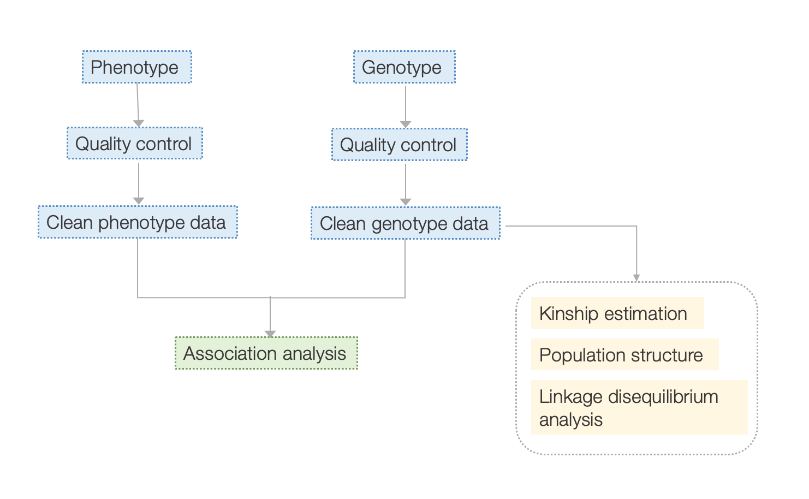
Nyocha data

Mgbe-ire ọrụ
Ọ gụnyere nyocha ndị a:
- Nyocha njikọ gbasara genome: LM, LMM, EMMAX, ụdị FASTLMM
- Nkọwa ọrụ nke mkpụrụ ndụ ihe nketa ahọpụtara
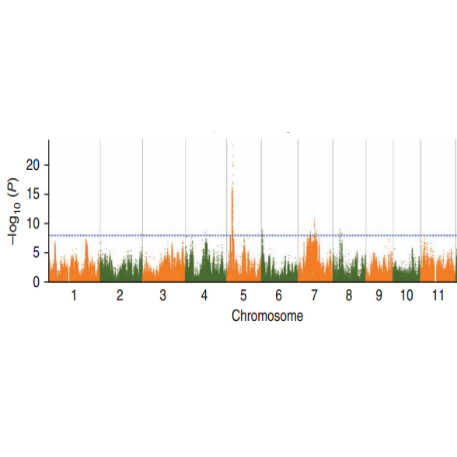
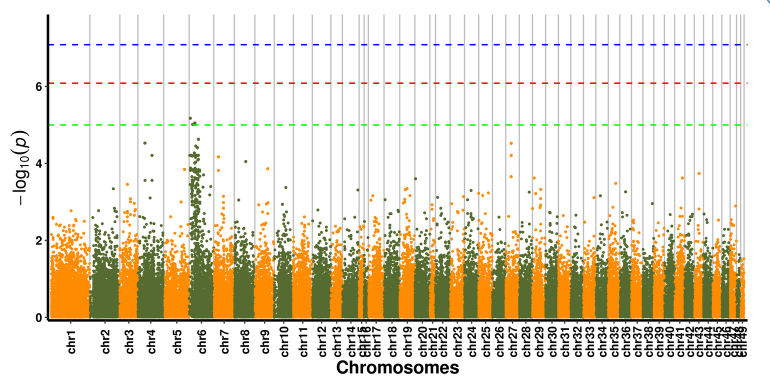
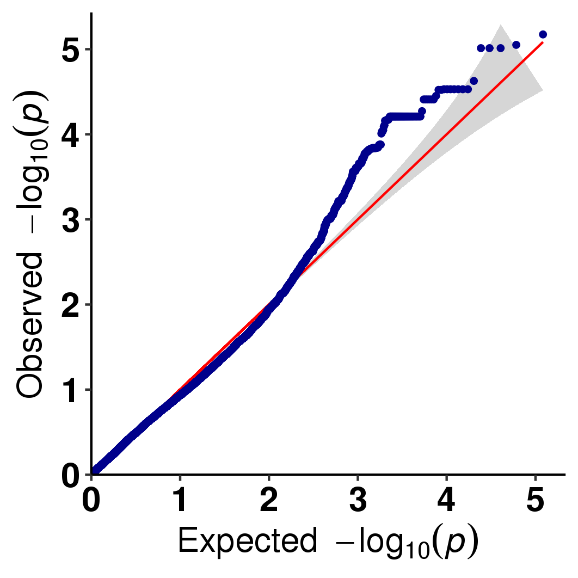
Nyocha mkpakọrịta nke SNP - atụmatụ Manhattan
Nyocha mkpakọrịta SNP-ụkpụrụ - atụmatụ QQ
Nyochaa ọganihu ndị ọrụ BMKGene's de GWAS na-akwado site na nchịkọta akwụkwọ ndị echekwabara:
Lv, L. et al. (2023) 'Nnyocha n'ime usoro mkpụrụ ndụ ihe nketa nke nnabata amonia na agụba clam Sinonovacula constricta site na ọmụmụ ihe ọmụmụ genome-wide',Aquaculture, 569, p. 739351. doi: 10.1016/J.AQUACULTURE.2023.739351.
Li, X. et al. (2022) 'Nyocha ọtụtụ-omics nke 398 foxtail millet accessions na-ekpughe mpaghara genomic metụtara ụlọ, àgwà metabolite, na mmetụta mgbochi mkpali',Osisi Molecular, 15 (8), p. 1367–1383. doi: 10.1016/j.molp.2022.07.003.
Li, J. et al. (2022) 'Genome-Wide Association Mapping nke Hulless Barely Phenotypes na Drought Environment',Oke na sayensị osisi, 13, p. 924892. doi: 10.3389/FPLS.2022.924892/BIBTEX.
Zhao, X. et al. (2021) 'GmST1, nke na-etinye koodu sulfotransferase, na-enye iguzogide nje mosaic soybean G2 na G3',Osisi, Cell & gburugburu, 44 (8), p. 2777–2792. doi: 10.1111/PCE.14066.