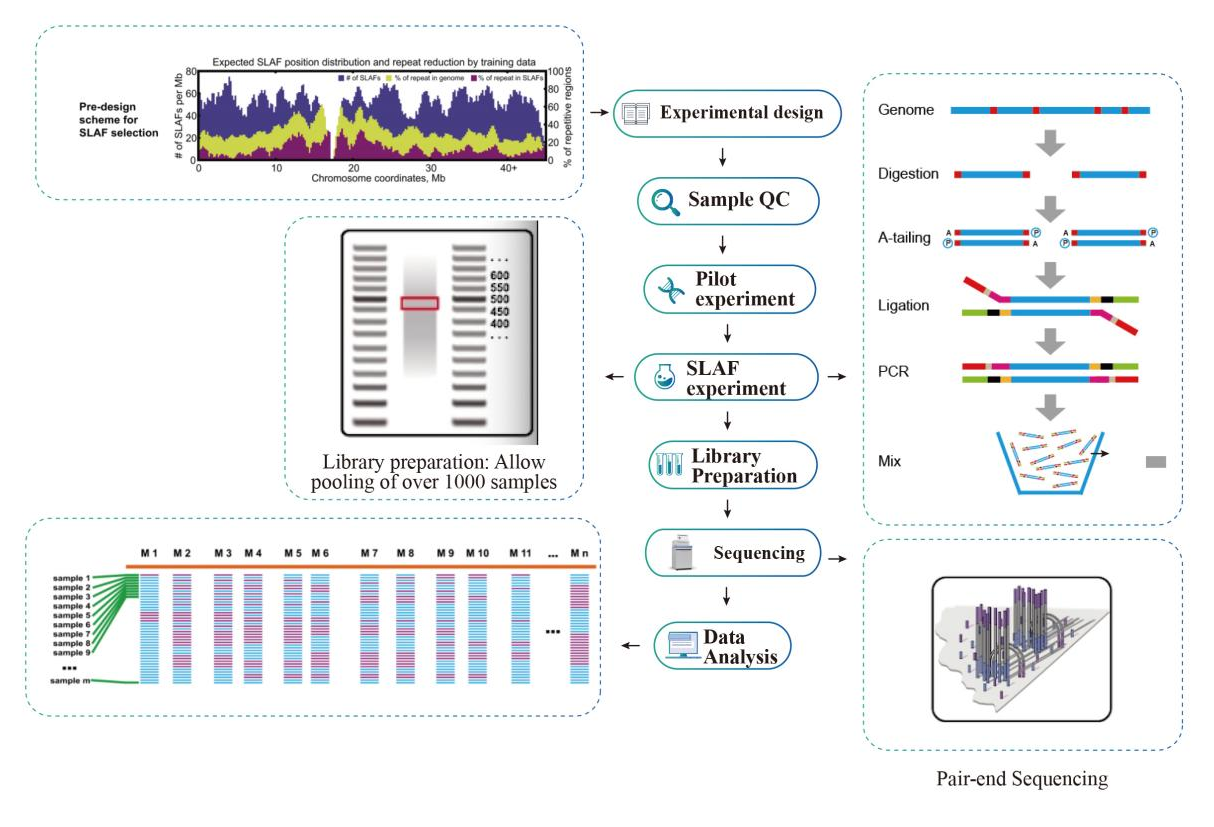
ʻO ke kaʻina o nā ʻāpana ʻāpana kikoʻī i hoʻonui ʻia (SLAF-Seq)
Nā hiʻohiʻona lawelawe
● Ke kaʻina ʻana ma NovaSeq me PE150.
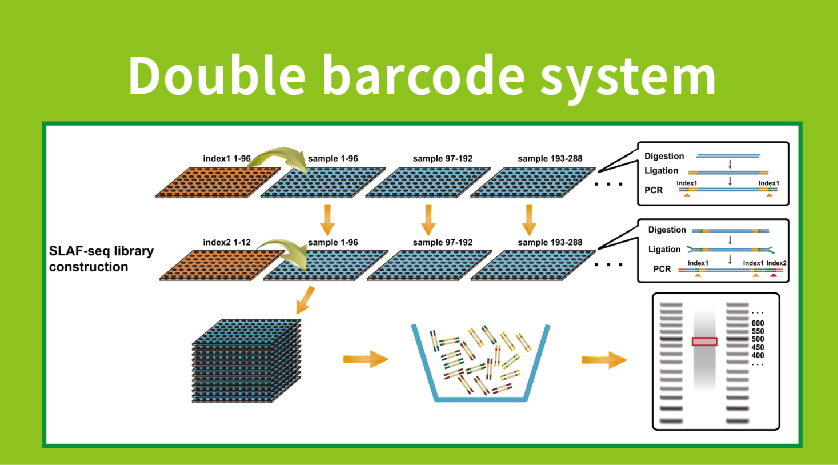
● Ka hoʻomākaukau ʻana o ka hale waihona puke me ka barcode pālua, hiki ke hoʻohui ʻia ma luna o 1000 mau laʻana.
● Hiki ke hoʻohana ʻia kēia ʻenehana me kahi genome kuhikuhi a i ʻole, me nā paipu bioinformatic like ʻole no kēlā me kēia hihia:
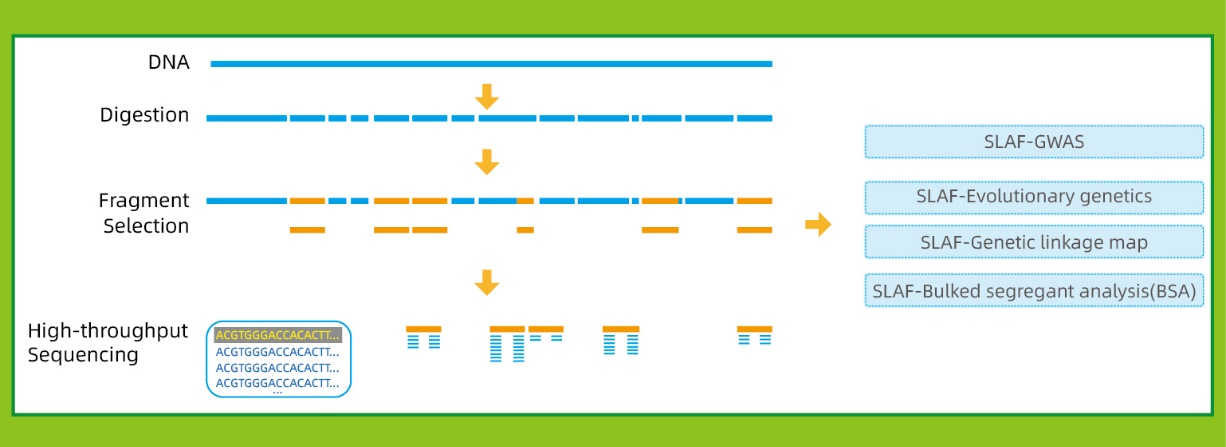
Me ka genome kuhikuhi: SNP a me InDel loaʻa
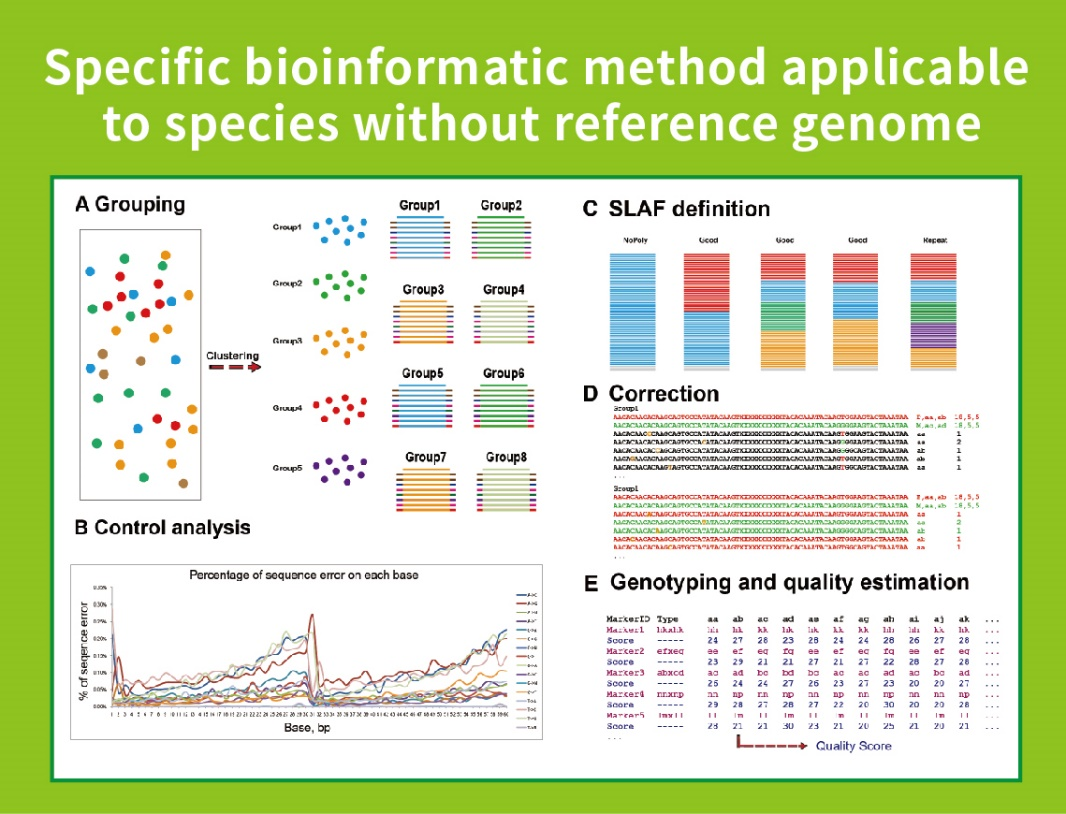
Me ka ʻole o ka genome hōʻike: hōʻuluʻulu laʻana a me ka ʻike SNP
● Ma kama ka silikoʻO ka pae mua o ka hoʻolālā e nānā ʻia nā hui ʻana o nā enzyme palena nui e ʻike i nā mea e hoʻopuka i ka hāʻawi like ʻana o nā hōʻailona SLAF ma ka genome.
● I ka wā o ka hoʻokolohua mua, ua hoʻāʻo ʻia ʻekolu hui ʻana o ka enzyme i 3 mau laʻana no ka hoʻohua ʻana i nā hale waihona puke 9 SLAF, a hoʻohana ʻia kēia ʻike e koho i ka hui ʻana o ka enzyme restriction maikaʻi loa no ka papahana.
Pono lawelawe
●ʻIke ʻia ʻo High Genetic Marker Discovery: ʻO ka hoʻohui ʻana i kahi ʻōnaehana barcode pālua kiʻekiʻe e hiki ai i ka hoʻonohonoho like ʻana o nā heluna nui, a ʻo ka hoʻonui ʻana i ka locus-specific e hoʻonui i ka pono, e hōʻoia ana i nā helu helu e hoʻokō i nā koi like ʻole o nā nīnau noiʻi like ʻole.
● Ke hilinaʻi haʻahaʻa i ka Genome: Hiki ke hoʻohana ʻia i nā ʻano me ka genome kuhikuhi a i ʻole.
●Hoʻolālā Papahana maʻalahi: Hoʻokahi-enzyme, dual-enzyme, multi-enzyme digestion, a me nā ʻano enzyme like ʻole hiki ke koho ʻia no ka hoʻokō ʻana i nā pahuhopu noiʻi ʻokoʻa. ʻO kama ka silikoHana ʻia ka hoʻolālā mua e hōʻoia i kahi hoʻolālā enzyme maikaʻi loa.
● Kiʻekiʻe Efficiency ma Enzymatic Digestion: Ka hana ana ama ka silikoʻO ka hoʻolālā mua a me ka hoʻokolohua mua i hōʻoia i ka hoʻolālā maikaʻi loa me ka hāʻawi like ʻana i nā huaʻōlelo SLAF ma ka chromosome (1 SLAF tag/4Kb) a me ka hōʻemi ʻana i ke kaʻina hana hou (<5%).
●ʻIke Nui: Hāʻawi kā mākou hui i ka waiwai o ka ʻike i kēlā me kēia papahana, me kahi moʻolelo o ka pani ʻana ma luna o 5000 mau papahana SLAF-Seq ma nā haneli haneli, me nā mea kanu, nā mammals, nā manu, nā inike, a me nā mea ola wai.
● Hoʻolālā ʻia ʻo Bioinformatic Workflow: Ua hoʻomohala ʻo BMKGENE i kahi kahe hana bioinformatic i hoʻohui ʻia no SLAF-Seq e hōʻoia i ka hilinaʻi a me ka pololei o ka hopena hope.
Nā kikoʻī lawelawe
| ʻAno kālailai | Paipai ʻia ka heluna kanaka | Hoʻolālā kaʻina | |
| ʻO ka hohonu o ka hoʻonohonoho ʻana i ka hōʻailona | Helu helu | ||
| Nā palapala ʻāina Genetic | 2 makua a >150 keiki | Nā mākua: 20x WGS Nā hua: 10x | Nui genome: <400 Mb: Manaʻo ʻia ʻo WGS <1Gb: 100K mau hōʻailona 1-2Gb:: 200K mau hōʻailona >2Gb: 300K mau hōʻailona Max 500k mau hōʻailona |
| Nā haʻawina ʻo Genome-Wide Association (GWAS) | ≥200 mau laʻana | 10x | |
| Genetic Evolution | ≥30 laʻana, me> 10 laʻana mai kēlā me kēia subgroup | 10x | |
Nā Koina lawelawe
Hoʻohui ≥ 5 ng/µL
Ka huina nui ≥ 80 ng
Nanodrop OD260/280=1.6-2.5
ʻO Agarose gel: ʻaʻohe a i ʻole ka hoʻohaʻahaʻa ʻana a i ʻole ka hoʻohaumia ʻana
Manaʻo ʻia ka hāʻawi laʻana
ʻO ka pahu: 2 ml centrifuge tube
(No ka hapa nui o nā laʻana, paipai mākou ʻaʻole e mālama i loko o ka ethanol)
Laʻana hōʻailona: Pono e hōʻailona ʻia nā laʻana a e like me ka palapala ʻike hāpana i waiho ʻia.
Hoʻouna: Ka hau maloʻo: Pono e hoʻopili mua ʻia nā laʻana i loko o nā ʻeke a kanu ʻia i loko o ka hau maloʻo.
Hana Hana Hana






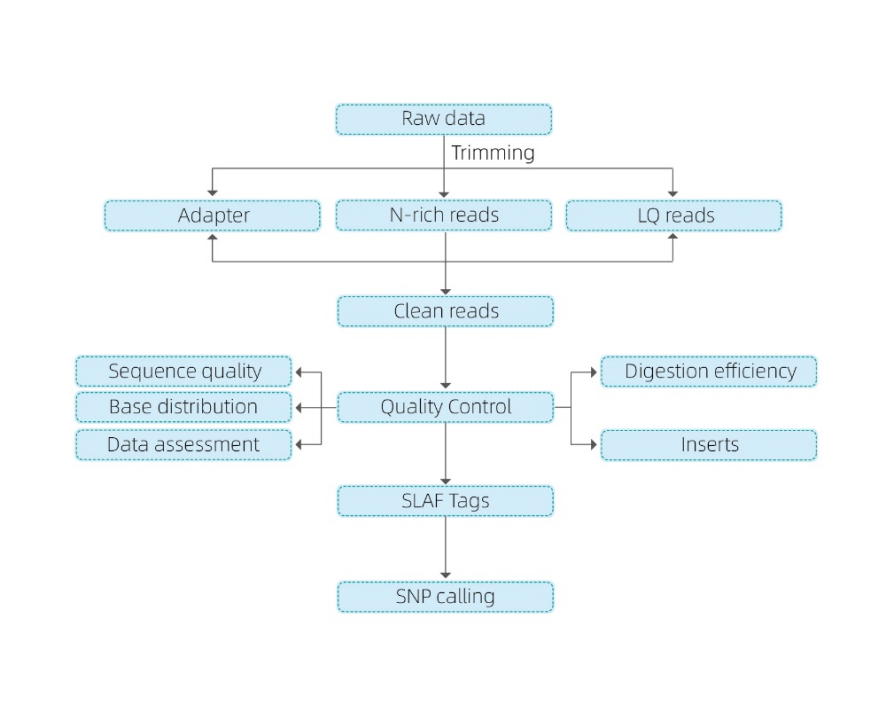
Laʻana QC
Hoʻokolohua hoʻokele
SLAF-hoʻokolohua
Hoʻomākaukau hale waihona puke
Ke kaʻina ʻana
ʻIkepili ʻikepili
Mahope o ke kuai ana
 Loaʻa i kēia ka nānā ʻana:
Loaʻa i kēia ka nānā ʻana:
- Ka helu helu QC
- Hoʻomohala ʻana i ka hōʻailona SLAF
Ka palapala ʻāina no ka kuhikuhi ʻana i ka genome
Me ka ʻole o ka genome kuhikuhi: clustering
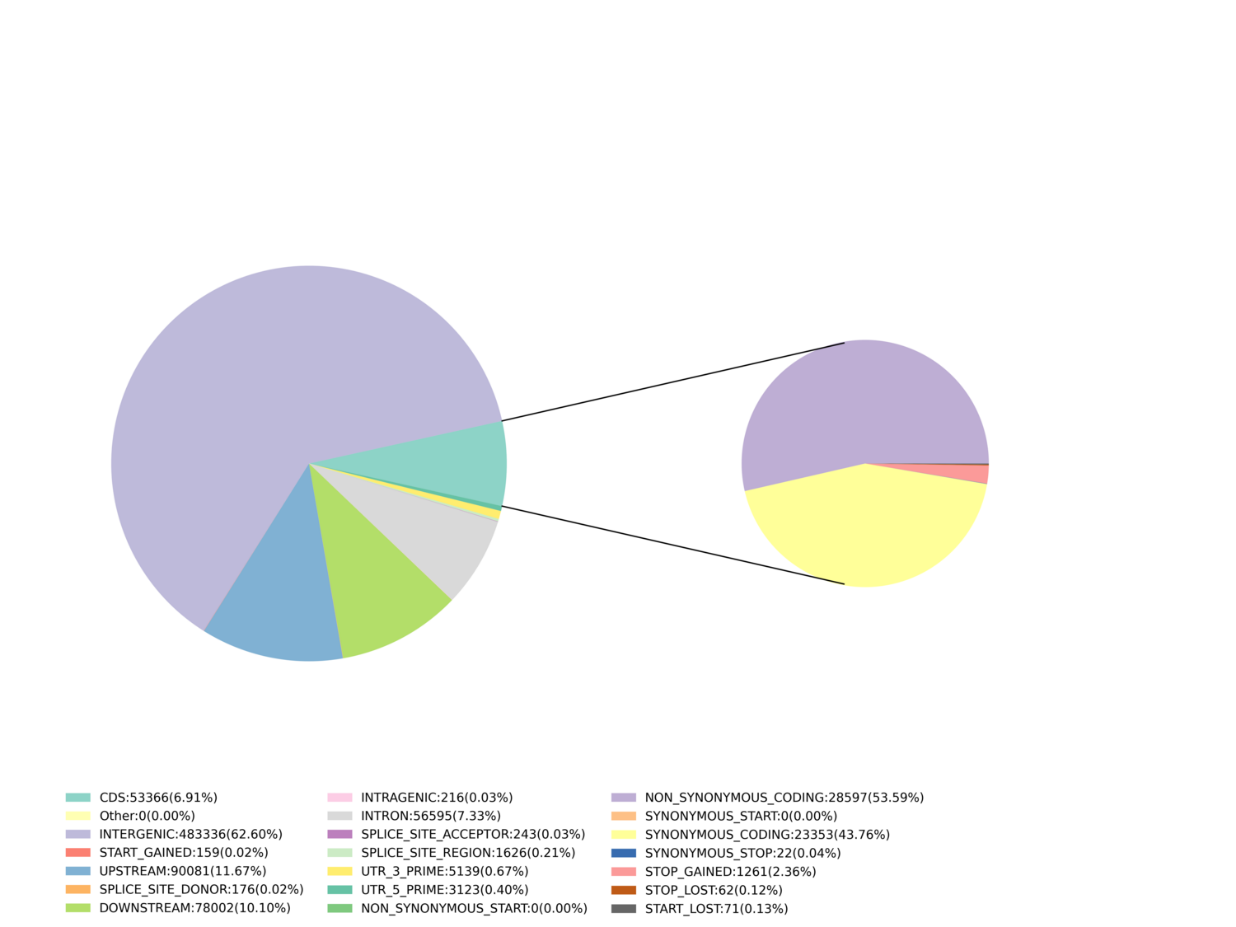
- Ka nānā 'ana i nā hua'ōlelo SLAF.: nā helu, ka hā'awi 'ana i ka genome
- ʻIke ʻia ka mākaʻikaʻi: SNP, InDel, SNV, kāhea CV a me ka hōʻike
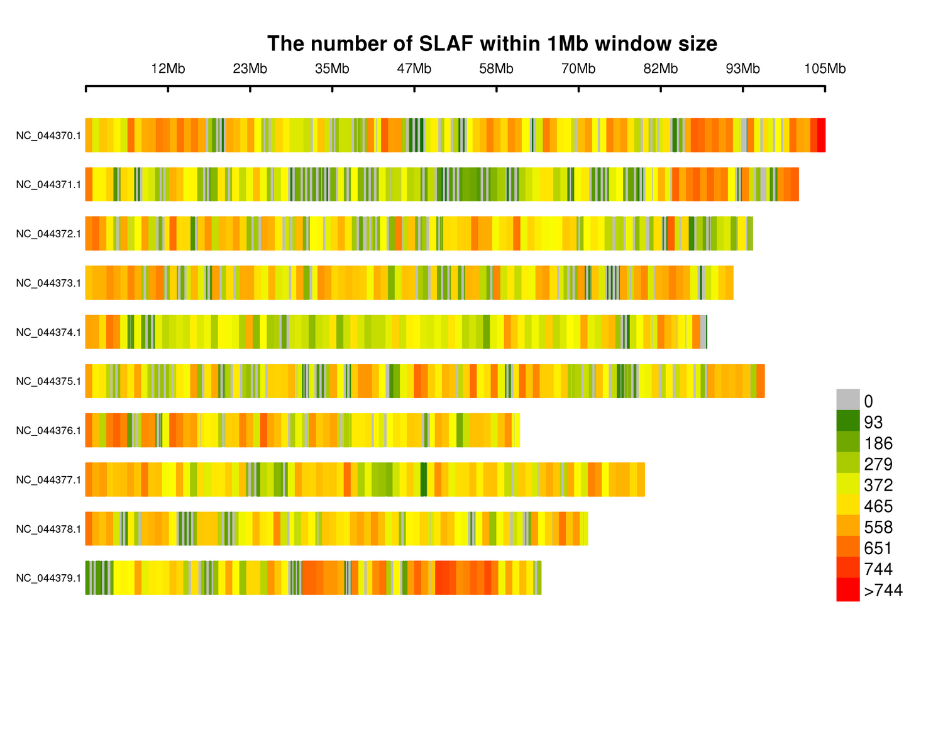
Ka mahele ʻana o nā hōʻailona SLAF ma nā chromosomes:
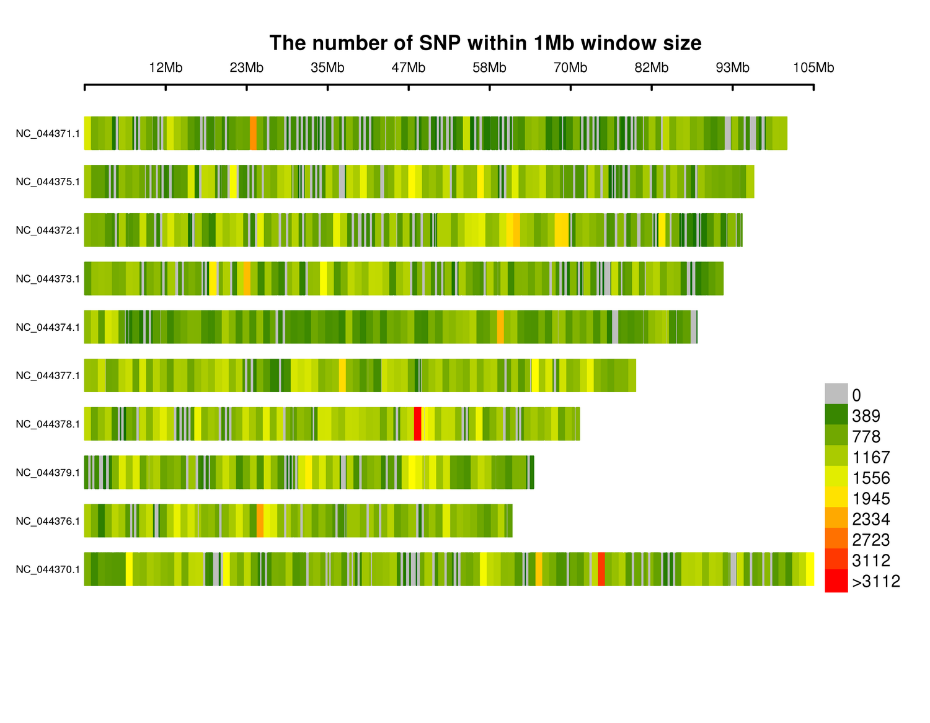
Ka māhele ʻana o nā SNP ma nā chromosomes:
| Makahiki | Nupepa | IF | Poʻo inoa | Nā noi |
| 2022 | Nā kamaʻilio kūlohelohe | 17.694 | Genomic kumu o ka giga-chromosomes a me ka giga-genome o ka laau peony Paeonia ostii | SLAF-GWAS |
| 2015 | Phytologist hou | 7.433 | Hoʻokumu nā kapuaʻi home i nā wahi genomic koʻikoʻi agronomic i soybeans | SLAF-GWAS |
| 2022 | Nūpepa no ka noiʻi kiʻekiʻe | 12.822 | ʻO ka hoʻokomo ʻana i ka genome-ākea o Gossypium barbadense i loko o G. hirsutum hōʻike i ka loci maikaʻi loa no ka hoʻomaikaʻi like ʻana o ka maikaʻi o ka pulupulu a me ka hua mau ano | SLAF-Evolutionary genetics |
| 2019 | Mea Molekula | 10.81 | Hōʻike ka heluna helu kanaka a me ka hui ʻo De Novo i ke kumu o ka weedy ʻO Laiki ma ke ʻano he pāʻani Evolutionary | SLAF-Evolutionary genetics |
| 2019 | Nature Genetics | 31.616 | ʻO ke kaʻina genome a me ka ʻokoʻa genetic o ka carp maʻamau, Cyprinus carpio | Palapala SLAF-Loina |
| 2014 | Nature Genetics | 25.455 | Hāʻawi ka genome o ka pīnī i kanu ʻia i ka ʻike i nā karyotypes legume, polyploid ka hooulu ana a me ka hoohana ana i na mea kanu. | Palapala SLAF-Loina |
| 2022 | Mea Puke Biotechnology Journal | 9.803 | Hōʻike ka ʻike ʻana o ST1 i kahi koho e pili ana i ka hitchhiking o ka morphology hua a me ka aila i loko o ka hana ʻana i ka soya | Hoʻomohala SLAF-Marker |
| 2022 | Nūpepa International o nā ʻepekema Molecular | 6.208 | ʻIke a me ka hoʻomohala ʻana i ka hōʻailona DNA no kahi Wheat-Leymus mollis 2Ns (2D) Hoʻololi ʻo Disomic Chromosome | Hoʻomohala SLAF-Marker |
| Makahiki | Nupepa | IF | Poʻo inoa | Nā noi |
| 2023 | Nā palena i ka ʻepekema mea kanu | 6.735 | ʻO ka palapala palapala QTL a me ka nānā ʻana i ka transcriptome o ke kō i ka wā o ka hua o Pyrus pyrifolia | Palapala ʻāina Genetic |
| 2022 | Mea Puke Biotechnology Journal | 8.154 | Hōʻike ka ʻike ʻana o ST1 i kahi koho e pili ana i ka hitchhiking o ka morphology hua a me ka ʻaila ʻaila i ka wā o ka home soybean.
| Kāhea ʻia ʻo SNP |
| 2022 | Nā palena i ka ʻepekema mea kanu | 6.623 | Genome-Wide Association Mapping of Hulless Barely Phenotypes in Drought Environment.
| GWAS |