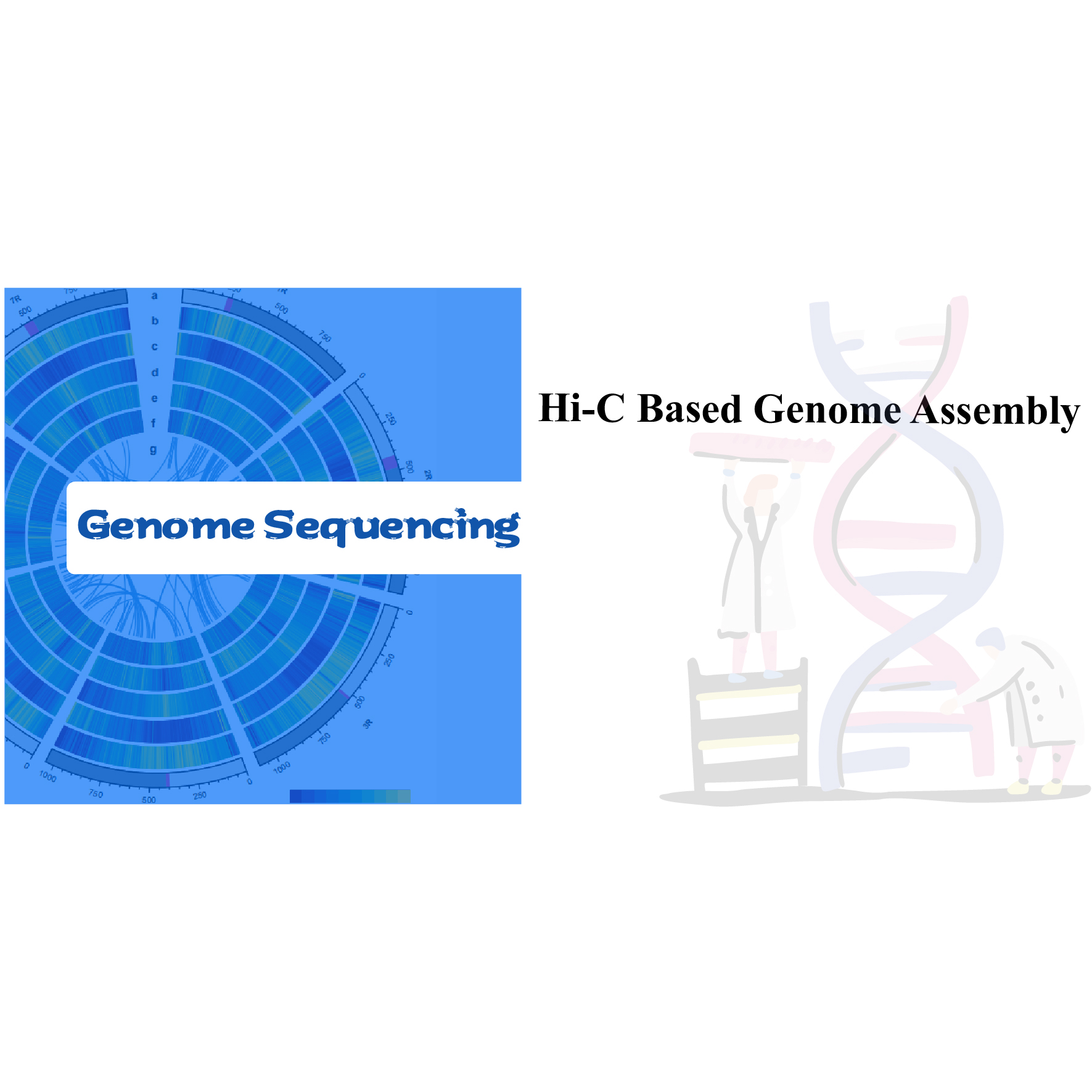
Mea kanu/holoholona holoʻokoʻa Genome Sequencing
Nā hiʻohiʻona lawelawe
● Hiki ke hoʻomākaukau ʻia ka waihona puke maʻamau a i ʻole PCR-free
● Loaʻa i loko o 4 sequencing platform: Illumina NovaSeq, MGI T7, Nanopore Promethion P48, a i ʻole PacBio Revio.
● ʻIke ʻia ka ʻike bioinformatic i ka ʻike ʻana i nā ʻano like ʻole: SNP, InDel, SV a me CNV
Pono lawelawe
●ʻIke Nui a me nā moʻolelo hoʻolaha: ʻO ka ʻike i hōʻiliʻili ʻia i ka genome sequencing no ʻoi aku ma mua o 1000 mau ʻano i hoʻopuka ʻia ma luna o 1000 mau hihia i paʻi ʻia me kahi kumu kumulative hopena ʻoi aku ma mua o 5000.
●ʻIkepili Bioinformatics piha: Me ka hoʻololi ʻana i ke kāhea ʻana a me ka hōʻike hana.
● Kākoʻo ma hope o ke kūʻai aku:ʻO kā mākou kūpaʻa e ʻoi aku ma mua o ka hoʻopau ʻana o ka papahana me kahi 3 mau mahina ma hope o ke kūʻai aku ʻana. I kēia manawa, hāʻawi mākou i ka hahai ʻana o ka papahana, kōkua hoʻoponopono, a me nā hālāwai Q&A e hoʻoponopono i nā nīnau pili i nā hopena.
●Hōʻikeʻike piha: Hoʻohana mākou i nā ʻikepili he nui no ka hōʻike ʻana i nā genes me nā ʻano like ʻole i ʻike ʻia a hana i ka loiloi hoʻonui kūpono, e hāʻawi ana i nā ʻike no nā papahana noiʻi lehulehu.
Nā kikoʻī lawelawe
| Nā ʻano like ʻole e ʻike ʻia | Hoʻolālā kaʻina | Paipai ʻia ka hohonu |
| SNP a me InDel | ʻO Illumina NovaSeq PE150 a i ʻole MGI T7 | 10x |
| SV a me CNV (emi pololei) | 30x | |
| SV a me CNV (ʻoi aku ka pololei) | Nanopore Prom P48 | 20x |
| Nā SNP, Indels, SV a me CNV | PacBio Revio | 10x |
Nā Koina Laʻana
| Tissue a i ʻole nā ʻakika nucleic i unuhi ʻia | Illumina/MGI | Nanopore | PacBio
| ||
| Holoholona Viscera | 0.5-1 g | ≥ 3.5 g
| ≥ 3.5 g
| ||
| ʻAiʻa holoholona | ≥ 5 g
| ≥ 5 g
| |||
| Koko Māmā | 1.5 mL | ≥ 0.5 mL
| ≥ 5 mL
| ||
| Koko moa/I'a | ≥ 0.1 mL
| ≥ 0.5 mL
| |||
| Mea kanu- Lau Hou | 1-2 g | ≥ 2 g
| ≥ 5 g
| ||
| Nā Pūnaehana Moʻomeheu |
| ≥ 1x107
| ≥ 1x108
| ||
| ʻO ka ʻiʻo palupalu nā pepeke/Kekahi | 0.5-1 g | ≥ 1 g
| ≥ 3 g
| ||
| DNA unuhi ʻia
| Hoʻopaʻa ʻia: ≥ 1 ng/ µL Ka nui: ≥ 30 ng Ua kaupalena ʻia a ʻaʻole hoʻohaʻahaʻa a hoʻohaumia paha
| Hoʻopaʻa ʻana Ka nui
OD260/280
OD260/230
Ua kaupalena ʻia a ʻaʻole hoʻohaʻahaʻa a hoʻohaumia paha
| ≥ 40 ng/ µL 4 µg/ke kahe wai/lapana
1.7-2.2
≥1.5 | Hoʻopaʻa ʻana Ka nui
OD260/280
OD260/230
Ua kaupalena ʻia a ʻaʻole hoʻohaʻahaʻa a hoʻohaumia paha | ≥ 50 ng/ µL 10 µg / kahe kahe / hāpana
1.7-2.2
1.8-2.5 |
| Hoʻomākaukau hale waihona puke ʻole PCR: Hoʻohui ≥ 40 ng/ µL Ka nui≥ 500 ng | |||||
Holo Hana Hana

Hāʻawi laʻana

Ka unuhi ʻana o DNA

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili
Hāʻawi ʻikepili
Loaʻa i kēia ka nānā ʻana:
- Ka Mana Mana Pono
- Heluhelu o ka hoʻolikelike ʻana i ka genome kuhikuhi
- ʻIke ʻokoʻa: SNP, InDel, SV a me CNV
- Hōʻike kikoʻī o nā ʻano like ʻole
Ka Ikepili o ka alignment to reference genome - sequencing depth distribution
Kāhea ʻia ʻo SNP ma waena o nā laʻana he nui
Hōʻike InDel - nā ʻikepili o ka lōʻihi o InDel ma ka ʻāina CDS a me ka ʻāpana genome-ākea
Hoʻolaha ʻokoʻa ma waena o ka genome - Circos plot
ʻO ka hōʻike kikoʻī o nā genes me nā ʻano like ʻole - Gene Ontology
Chai, Q. et al. (2023) 'A glutathione S-transferase GhTT19 e hoʻoholo i ka pua puaʻa pua ma o ka hoʻoponopono ʻana i ka hōʻiliʻili anthocyanin i loko o ka pulupulu', Plant Biotechnology Journal, 21(2), p. 433. doi: 10.1111/PBI.13965.
Cheng, H. et al. (2023) Hāʻawi ʻo 'Chromosome-level wild Hevea brasiliensis genome i nā mea hana hou no ka hānau ʻana i kōkua ʻia i ka genomic a me ka loci waiwai e hoʻonui ai i ka hoʻohua ʻana o ka lāʻau', Plant Biotechnology Journal, 21(5), pp. 1058–1072. doi: 10.1111/PBI.14018.
Li, A. et al. (2021) 'Hāʻawi ʻo Genome of the estuarine oyster i nā ʻike i ka hopena o ke aniau a me ka plasticity adaptive', Communications Biology 2021 4:1, 4(1), pp. 1–12. doi: 10.1038/s42003-021-02823-6.
Zeng, T. et al. (2022) 'O ka loiloi o ka genome a me ka methylation hoʻololi i nā moa ʻōiwi Kina i ka manawa e hāʻawi i ka ʻike i ka mālama ʻana i nā ʻano mea', Communications Biology, 5(1), pp. 1–12. doi: 10.1038/s42003-022-03907-7.