PacBio 2+3 Hoʻonā mRNA piha piha
Nā hiʻohiʻona
● Haʻawina hoʻolālā:
Hoʻopili ʻia ka laʻana me PacBio e ʻike i nā isoforms transcript
E hoʻokaʻawale i nā laʻana (nā hoʻopiʻi a me nā kūlana e hoʻāʻo ʻia) i hoʻonohonoho ʻia meNGS e helu i ka ʻōlelo transcript
● PacBio sequencing in CCS mode, generating HiFi reads
● Ke kaʻina ʻana o nā moʻolelo holoʻokoʻa
● ʻAʻole pono ka ʻikepili i kahi genome kuhikuhi; akā, hiki ke hoʻohana ʻia
● ʻAʻole wale ka hōʻike ʻana ma ka gene a me ka isoform-level ka nānā ʻana i ka bioinformatic akā ʻo ka nānā ʻana i ka lncRNA, ka hui pū ʻana, ka poly-adenylation, a me ka hoʻolālā gene.
Nā pono
● Pono Kiʻekiʻe: Heluhelu ʻo HiFi me ka pololei >99.9% (Q30), e like me NGS
● Nānā Hoʻopili ʻokoʻa: ʻO ka hoʻonohonoho ʻana i nā transcript āpau e hiki ai ke ʻike isoform a me ke ʻano.
● Hui o PacBio a me NGS ikaika: hiki i ka helu ʻana o ka ʻōlelo ma ka pae isoform, wehe i ka loli i uhi ʻia i ka wā e nānā ana i ka hōʻike gene holoʻokoʻa.
● ʻIke Nui: me kahi moʻolelo o ka hoʻopau ʻana ma luna o 1100 PacBio piha transcriptome papahana a me ka hana ʻana ma luna o 2300 mau laʻana, lawe kā mākou hui i ka waiwai o ka ʻike i kēlā me kēia papahana.
● Kākoʻo ma hope o ke kūʻai aku: ʻo kā mākou kūpaʻa e ʻoi aku ma mua o ka hoʻopau ʻana o ka papahana me kahi manawa lawelawe 3 mahina ma hope o ke kūʻai aku. I kēia manawa, hāʻawi mākou i ka hahai ʻana o ka papahana, kōkua hoʻoponopono, a me nā hālāwai Q&A e hoʻoponopono i nā nīnau pili i nā hopena.
Nā Koina Laʻana a me ka Hoʻouna ʻana
| Hale Waihona Puke | Hoʻolālā kaʻina | Manaʻo ʻia ka ʻikepili | Ka Mana Mana |
| Hoʻonui ʻia ʻo PolyA mRNA CCS waihona | PacBio Sequel II PacBio Revio | 20/40 Gb 5/10 M CCS | Q30≥85% |
| Hoʻonui ʻia ʻo Poly A | Illumina PE150 | 6-10 Gb | Q30≥85% |
Nucleotides
|
| Conc.(ng/μl) | Ka nui (μg) | Maemae | Pono pono |
| Hale Waihona Puke Illumina | ≥ 10 | ≥ 0.2 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Hoʻopaʻa ʻia a ʻaʻole hoʻi i ka protein a i ʻole DNA contamination i hōʻike ʻia ma ka gel. | No nā mea kanu: RIN≥4.0; No nā holoholona: RIN≥4.5; 5.0≥28S/18S≥1.0; palena a ʻaʻole kiʻekiʻe kumu |
| hale waihona puke ʻo PacBio | ≥ 100 | ≥ 1.0 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Hoʻopaʻa ʻia a ʻaʻole hoʻi i ka protein a i ʻole DNA contamination i hōʻike ʻia ma ka gel. | Nā mea kanu: RIN≥7.5 Nā holoholona: RIN≥8.0 5.0≥28S/18S≥1.0; palena a ʻaʻole kiʻekiʻe kumu |
Manaʻo ʻia ka hāʻawi laʻana
Pahu: 2 ml centrifuge tube (ʻAʻole ʻōlelo ʻia ʻo Tin foil)
Laʻana lepili: Pūʻulu+hoʻopiʻi e laʻa me A1, A2, A3; B1, B2, B3.
Hoʻouna:
1. Hau maloo:Pono e hoʻopili ʻia nā laʻana i loko o nā ʻeke a kanu ʻia i loko o ka hau maloʻo.
2. Nā paipu RNAstable: Hiki ke hoʻomaloʻo ʻia nā laʻana RNA i loko o ka paipu hoʻopaʻa RNA (e laʻa me RNAstable®) a hoʻouna ʻia i loko o ka lumi wela.
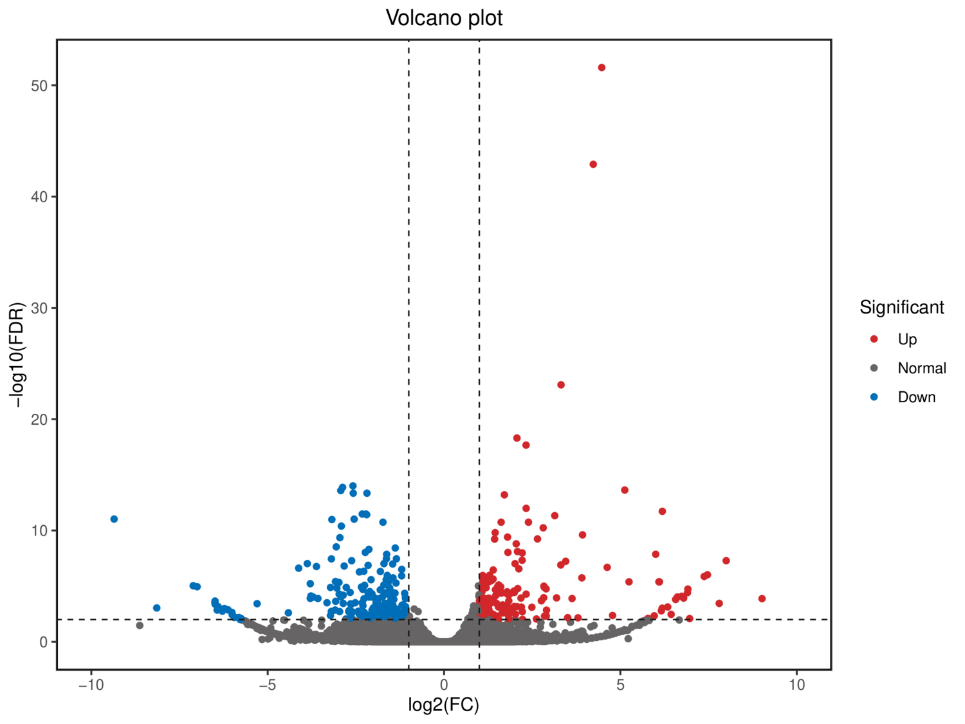
Loaʻa i kēia ka nānā ʻana:
Ka mana maikaʻi ʻikepili maka
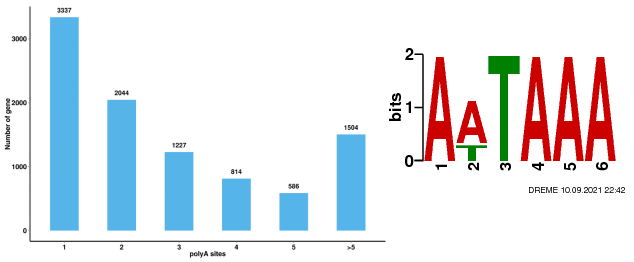
ʻIkepili Polyadenylation ʻē aʻe (APA)
Hoʻopili transcript analysis
ʻAno Hoʻopili Hoʻopili ʻana
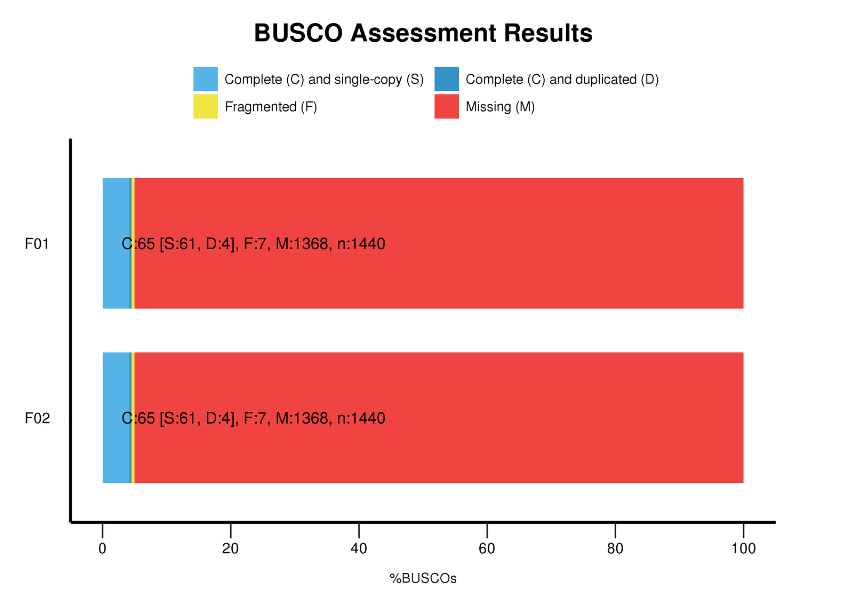
Ka nānā 'ana i nā Universal Single-Copy Orthologs (BUSCO).
ʻO ka loiloi transcript hou: wānana o nā kaʻina coding (CDS) a me ka hōʻike hana
Ka nānā ʻana o lncRNA: wānana o lncRNA a me nā pahuhopu
ʻIkepili MicroSatelite (SSR)
Ka nānā 'ana i nā palapala kope i hō'ike 'oko'a (DETs).
Ka nānā 'ana i nā Genes 'oko'a (DEGs).
ʻO ka hōʻike kikoʻī o nā DEG a me DET
ʻIkepili BUSCO
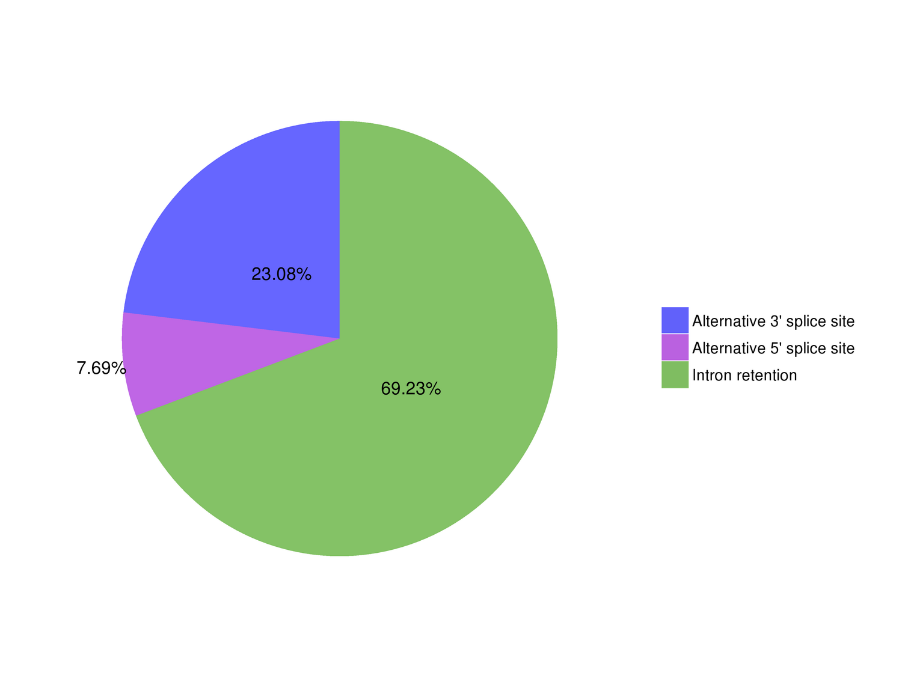
ʻAno Hoʻopili Hoʻopili ʻana
ʻIkepili Polyadenylation ʻē aʻe (APA)
Nā Genes Hōʻike ʻokoʻa (DEGs) a me nā Transcripts (DETs9 anlaysis
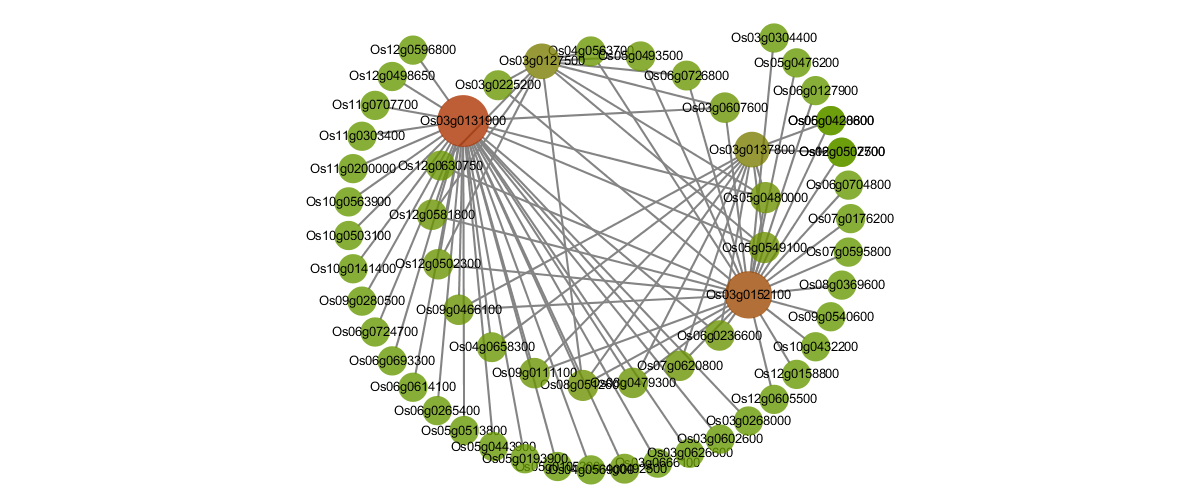
ʻO nā pūnaewele pilina protein-Protein o nā DET a me nā DEG
E ʻimi i nā holomua i hoʻomaʻamaʻa ʻia e BMKGene's PacBio 2+3 holo piha mRNA sequencing ma o kahi hōʻiliʻili curated o nā puke.
Chao, Q. et al. (2019) 'The developmental dynamics of the Populus stem transcriptome', Plant Biotechnology Journal, 17(1), pp. 206–219. doi: 10.1111/PBI.12958.
Deng, H. et al. (2022) 'Dynamic Changes in Ascorbic Acid Content during Fruit Development and Ripening of Actinidia latifolia (he Ascorbate-Rich Fruit Crop) and the Associated Molecular Mechanisms', International Journal of Molecular Sciences, 23(10), p. 5808. doi: 10.3390/IJMS23105808/S1.
Hua, X. et al. (2022) 'Wanaana kūpono o nā genes ala biosynthetic i komo i nā polyphyllins bioactive ma Paris polyphylla', Communications Biology 2022 5:1, 5(1), pp. 1–10. doi: 10.1038/s42003-022-03000-z.
Liu, M. et al. (2023) 'Hoʻohui ʻia ʻo PacBio Iso-Seq a me Illumina RNA-Seq Analysis of the Tuta absoluta (Meyrick) Transcriptome and Cytochrome P450 Genes', Insects, 14(4), p. 363. doi: 10.3390/INSECTS14040363/S1.
Wang, Lijun et al. (2019) 'He noiʻi o ka paʻakikī transcriptome me ka hoʻohana ʻana iā PacBio hoʻokahi-molele i ka manawa maoli i hui pū ʻia me ka Illumina RNA sequencing no ka ʻike maikaʻi ʻana i ka biosynthesis acid ricinoleic ma Ricinus communis', BMC Genomics, 20(1), pp. 1–17. doi: 10.1186 / S12864-019-5832-9 / NUI / 7.