Non-Reference based mRNA Sequencing-NGS
Nā hiʻohiʻona
● Paʻa i ka poly mRNA ma mua o ka hoʻomākaukau ʻana i ka waihona
● Kūʻokoʻa i kēlā me kēia genome kuhikuhi: ma muli o ka hui de novo o nā transcripts, e hana ana i kahi papa inoa o nā unigenes i hōʻike ʻia me nā ʻikepili he nui (NR, Swiss-Prot, COG, KOG, eggNOG, Pfam, GO, KEGG)
● Hoʻopili bioinformatic piha o ka hōʻike gene a me ke ʻano transcript
Pono lawelawe
●ʻIke Nui: me ka mooolelo o ka hana ana ma luna o 600,000 laʻana ma BMKGENE, e pili ana i nā ʻano laʻana like ʻole e like me nā moʻomeheu cell, nā ʻiʻo, a me nā wai kino, lawe mai kā mākou hui i ka waiwai o ka ʻike i kēlā me kēia papahana. Ua pani maikaʻi mākou ma luna o 100,000 mau papahana mRNA-Seq ma nā kikowaena noiʻi like ʻole.
●Hoʻomalu maikaʻi loa: hoʻokō mākou i nā wahi mana koʻikoʻi ma nā pae āpau, mai ka laʻana a me ka hoʻomākaukau ʻana i ka waihona a hiki i ka sequencing a me nā bioinformatics. Mālama kēia mākaʻikaʻi ʻana i ka hāʻawi ʻana i nā hopena kiʻekiʻe mau.
● Hōʻikeʻike piha: hoʻohana mākou i nā ʻikepili he nui e hoʻopuka pono i nā Genes Differentially Expressed (DEGs) a hana i ka loiloi hoʻonui e pili ana, e hāʻawi ana i nā ʻike e pili ana i nā kaʻina kelepona a me nā molekala e pili ana i ka pane transcriptome.
●Kākoʻo ma hope o ke kūʻai aku: ʻo kā mākou kūpaʻa e ʻoi aku ma mua o ka hoʻopau ʻana o ka papahana me kahi manawa lawelawe 3 mahina ma hope o ke kūʻai aku. I kēia manawa, hāʻawi mākou i ka hahai ʻana o ka papahana, kōkua hoʻoponopono, a me nā hālāwai Q&A e hoʻoponopono i nā nīnau pili i nā hopena.
Nā Koina Laʻana a me ka Hoʻouna ʻana
| Hale Waihona Puke | Hoʻolālā kaʻina | Manaʻo ʻia ka ʻikepili | Ka Mana Mana |
| Hoʻonui ʻia ʻo Poly A | Illumina PE150 DNSSEQ-T7 | 6-10 Gb | Q30≥85% |
Nā Koina Laʻana:
Nucleotides:
| Conc.(ng/μl) | Ka nui (μg) | Maemae | Pono pono |
| ≥ 10 | ≥ 0.2 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Hoʻopaʻa ʻia a ʻaʻole hoʻi i ka protein a i ʻole DNA contamination i hōʻike ʻia ma ka gel. | No nā mea kanu: RIN≥4.0; No nā holoholona: RIN≥4.5; 5.0≥28S/18S≥1.0; palena a ʻaʻole kiʻekiʻe kumu |
● Nā mea kanu:
Aʻa, ʻaʻa a i ʻole Petal: 450 mg
Lau a Hua paha: 300 mg
Hua: 1.2 g
● holoholona:
Puʻuwai a i ʻole ka ʻōpū: 300 mg
ʻO Viscera a i ʻole lolo: 240 mg
ʻiʻo: 450 mg
Iwi, lauoho a ʻili paha: 1g
● Nā ʻāʻī:
Nā Insect: 6g
Crustacea: 300 mg
● Ke koko holoʻokoʻa: 1 paipu
● Nā Pūnaewele: 106 pūnaewele
Manaʻo ʻia ka hāʻawi laʻana
Pahu: 2 ml centrifuge tube (ʻAʻole ʻōlelo ʻia ʻo Tin foil)
Laʻana lepili: Pūʻulu+hoʻopiʻi e laʻa me A1, A2, A3; B1, B2, B3.
Hoʻouna:
1. Ka hau maloo: Pono e hoʻopili ʻia nā laʻana i loko o nā ʻeke a kanu ʻia i loko o ka hau maloʻo.
2. Nā paipu RNAstable: Hiki ke hoʻomaloʻo ʻia nā laʻana RNA i loko o ka paipu hoʻopaʻa RNA (e laʻa me RNAstable®) a hoʻouna ʻia i loko o ka lumi wela.
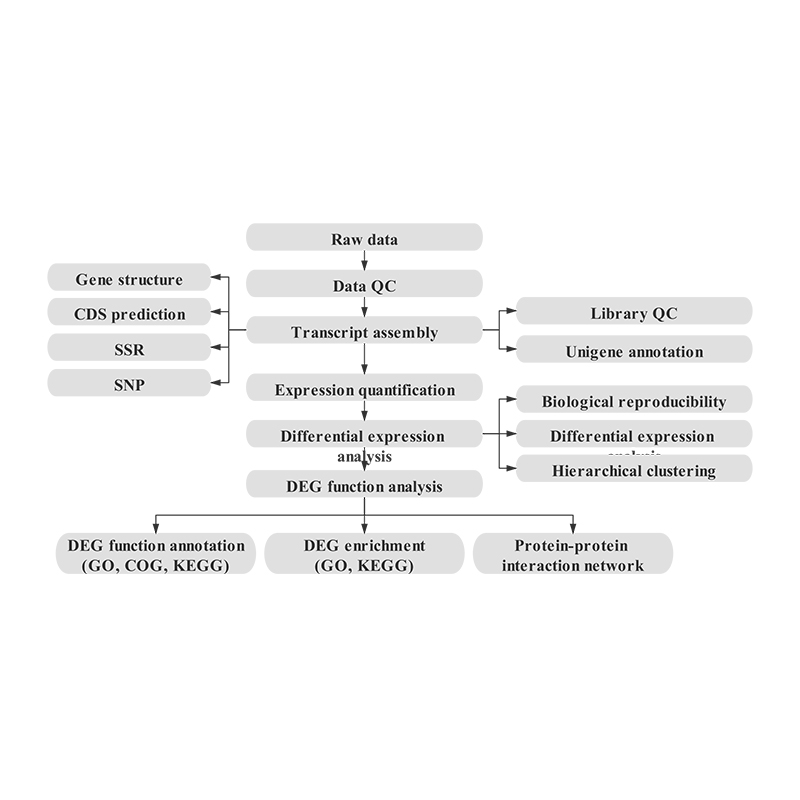
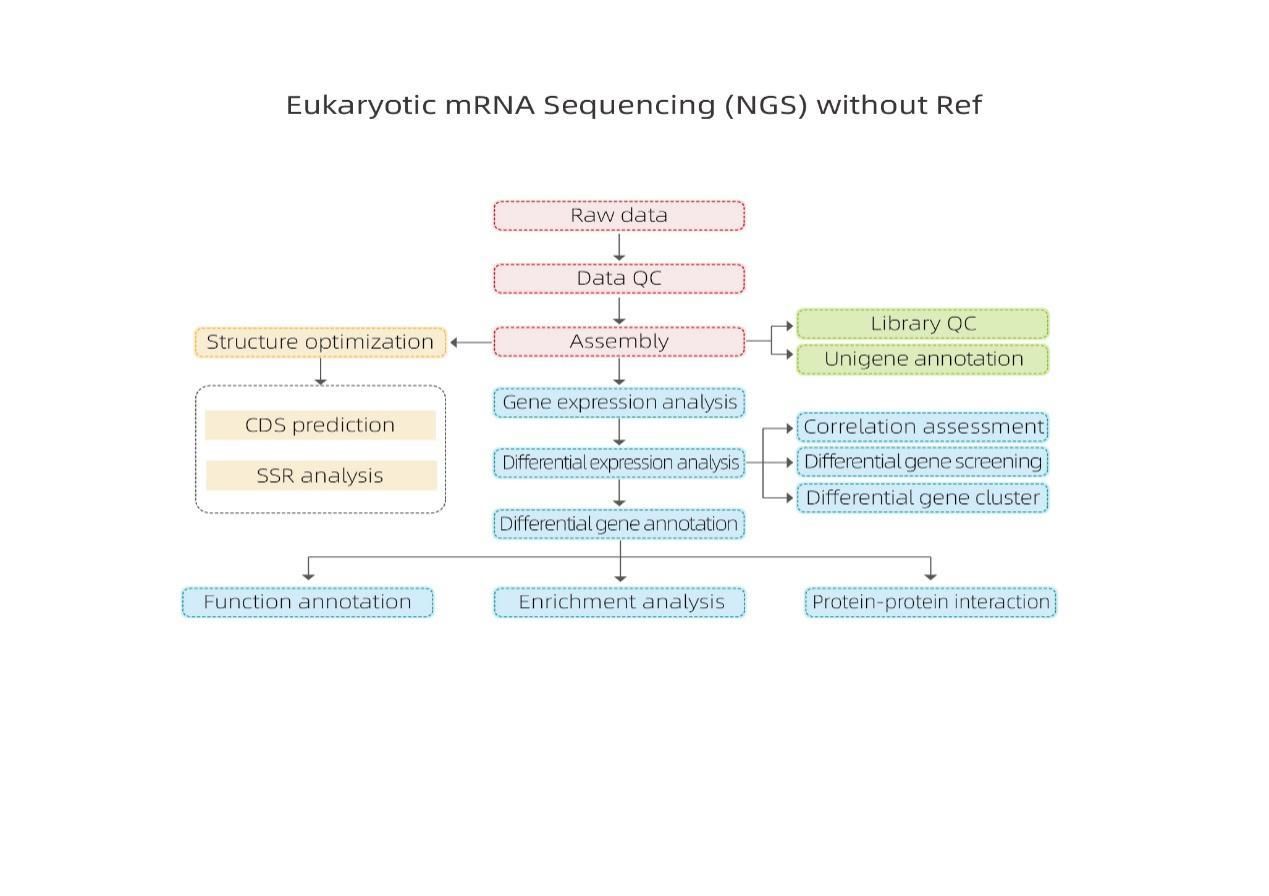
Holo Hana Hana

Hoʻolālā hoʻokolohua

Hāʻawi laʻana

RNA unuhi

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
Bioinformatics
ʻO ka hui transcriptome a me ke koho unigene
Hōʻike Unigene
Hoʻopili laʻana a me ka loiloi o nā hoʻopiʻi koʻikoʻi
Nā Genes Hōʻike ʻokoʻa (DEGs)
Hōʻike hana o nā DEG
Hoʻonui i ka hana o nā DEG
E ʻimi i nā holomua i hoʻolako ʻia e BMKGene's eukaryotic NGS mRNA sequencing services ma o ka hōʻiliʻili ʻana o nā puke.
Shen, F. et al. (2020) 'De novo transcriptome assembly and sex-biased gene expression in the gonads of Amur catfish (Silurus asotus)', Genomics, 112(3), pp. 2603–2614. doi: 10.1016/J.YGENO.2020.01.026.
Zhang, C. et al. (2016) 'Transcriptome analysis of sucrose metabolism i ka wā o ka ʻōpuʻu puʻupuʻu a me ka hoʻomohala ʻana i ka onion (Allium cepa L.)', Frontiers in Plant Science, 7 (September), p. 212763. doi: 10.3389/FPLS.2016.01425/BIBTEX.
Zhu, C. et al. (2017) 'De novo hui, hōʻike a me ka hōʻike no ka transcriptome o Sarcocheilichthys sinensis', PLoS ONE, 12 (2). doi: 10.1371/JOURNAL.PONE.0171966.
Zou, L. et al. (2021) Hāʻawi ka 'De novo transcriptome analysis i nā ʻike i ka hoʻomanawanui paʻakai o Podocarpus macrophyllus ma lalo o ke koʻikoʻi paʻakai', BMC Plant Biology, 21(1), pp. 1–17. doi: 10.1186 / S12870-021-03274-1 / NUI / 9.