ʻIkepili ʻAhahui Genome-wide
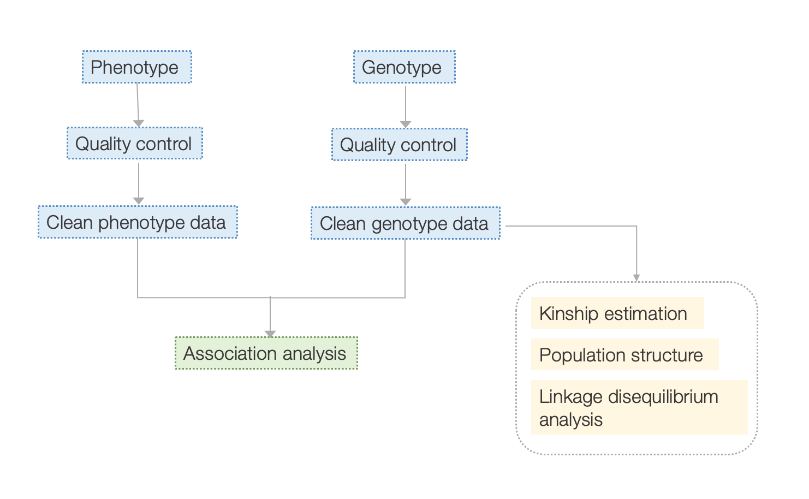
Kaʻina hana
Pono lawelawe
●ʻIke Nui a me nā moʻolelo hoʻolaha: me ka ʻike i hōʻiliʻili ʻia ma GWAS, ua hoʻopau ʻo BMKGene i nā haneli he nui o nā ʻano papahana i ka noiʻi ʻana o ka heluna kanaka GWAS, kōkua i nā mea noiʻi e hoʻopuka i nā ʻatikala ʻoi aku ma mua o 100, a ua hōʻea ka kumulative hopena i 500.
● Ka nānā 'ana i nā bioinformatics piha: Loaʻa i ka holo kaʻa hana ka nānā ʻana o ka hui SNP-trait, e hāʻawi ana i kahi pūʻulu o nā genes moho a me kā lākou palapala kikoʻī hana.
●ʻO ka hui bioinformatics akamai loa a me ka pōʻaiapili pōkole: me ka ʻike maikaʻi loa i ka loiloi genomics kiʻekiʻe, hāʻawi ka hui o BMKGene i nā loiloi piha me ka manawa hoʻololi wikiwiki.
●Kākoʻo ma hope o ke kūʻai ʻana:ʻO kā mākou kūpaʻa e ʻoi aku ma mua o ka hoʻopau ʻana o ka papahana me kahi 3 mau mahina ma hope o ke kūʻai aku ʻana. I kēia manawa, hāʻawi mākou i ka hahai ʻana o ka papahana, kōkua hoʻoponopono, a me nā hālāwai Q&A e hoʻoponopono i nā nīnau pili i nā hopena.
Nā kikoʻī lawelawe a me nā koi
| ʻAno kaʻina | Paipai ʻia ka heluna kanaka | Hoʻolālā kaʻina | Koi nucleotide |
| Ka Hoʻopili Genome holoʻokoʻa | 200 laʻana | 10x | Hoʻopaʻa ʻia: ≥ 1 ng/ µL Ka huina nui≥ 30ng Ua kaupalena ʻia a ʻaʻole hoʻohaʻahaʻa a hoʻohaumia paha |
| Māhele Kūikawā-Locus Amplified Fragment (SLAF) | Ka hohonu o ka tag: 10x Ka helu o nā hōʻailona: <400 Mb: Manaʻo ʻia ʻo WGS <1Gb: 100K mau hōʻailona 1Gb > 2Gb: 300K mau hōʻailona Max 500k mau hōʻailona | Hoʻohui ≥ 5 ng/µL Ka huina nui ≥ 80 ng Nanodrop OD260/280=1.6-2.5 ʻO Agarose gel: ʻaʻohe a i ʻole ka hoʻohaʻahaʻa ʻana a i ʻole ka hoʻohaumia ʻana
|
Koho Mea



Nā ʻano like ʻole, subspecies, landraces/genebanks/mixed family/wild resources
ʻO nā ʻano like ʻole, subspecies, landraces
ʻOhana hapa-sib/ʻohana-sib piha/ ʻohana ʻāhiu
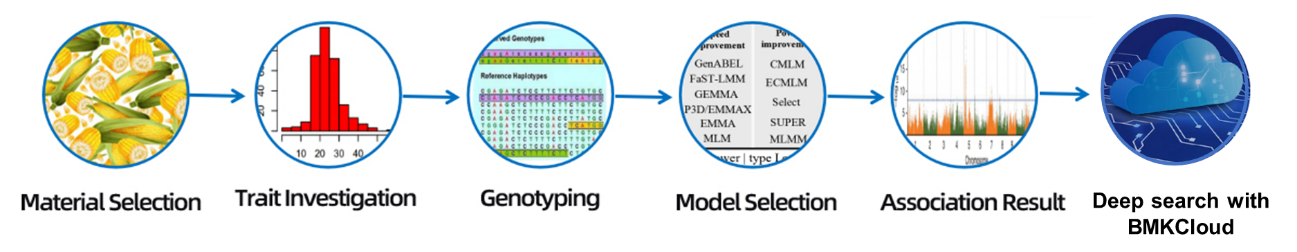
Holo Hana Hana

Hoʻolālā hoʻokolohua

Hāʻawi laʻana

RNA unuhi

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
Loaʻa i kēia ka nānā ʻana:
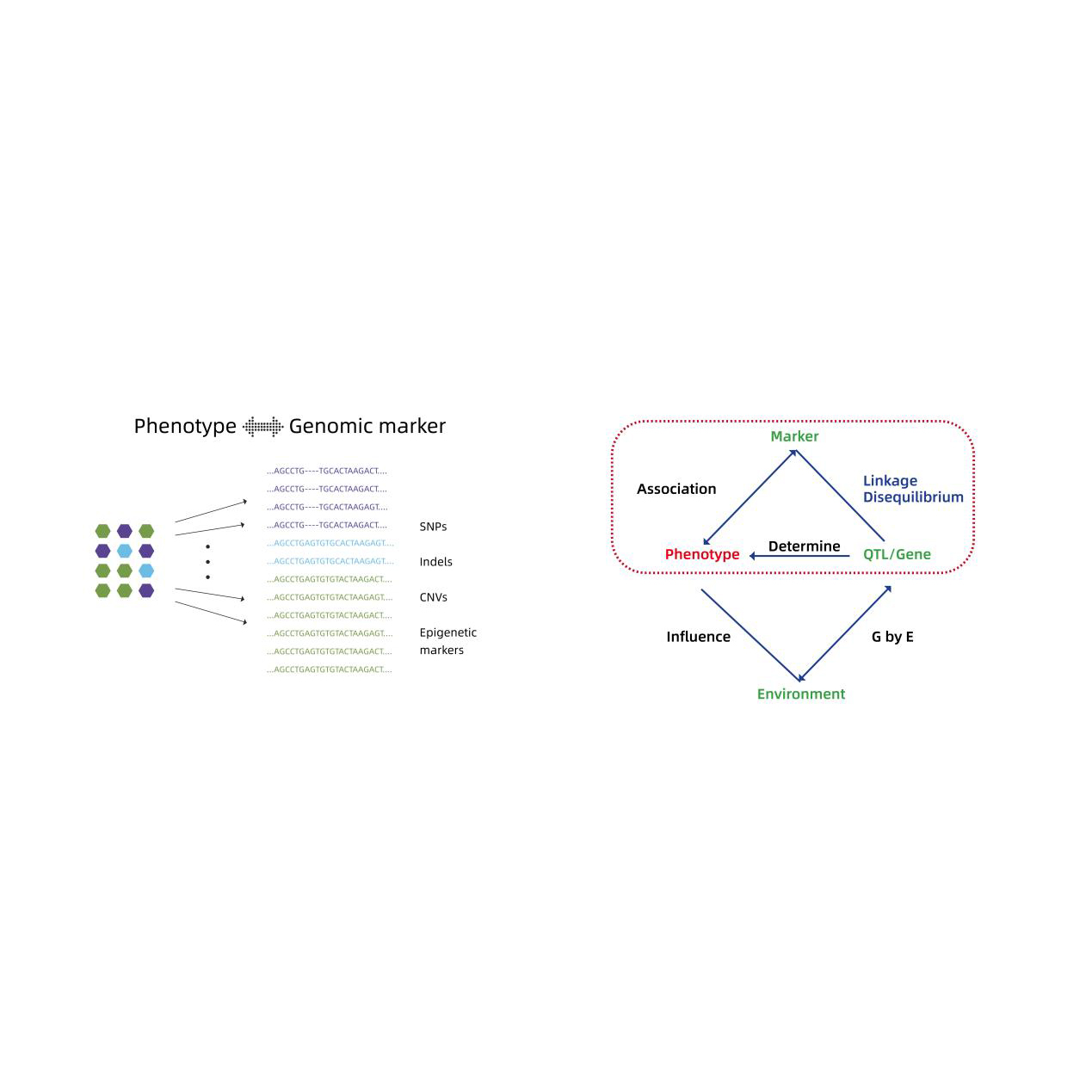
- Ka nānā 'ana i ka hui pū 'ana o ka genome: LM, LMM, EMMAX, FASTLMM model
- Hōʻike kikoʻī o nā genes moho
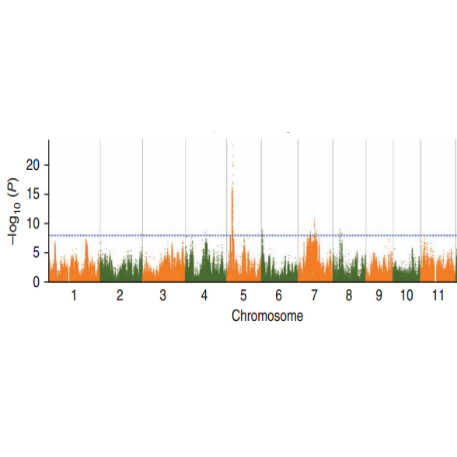
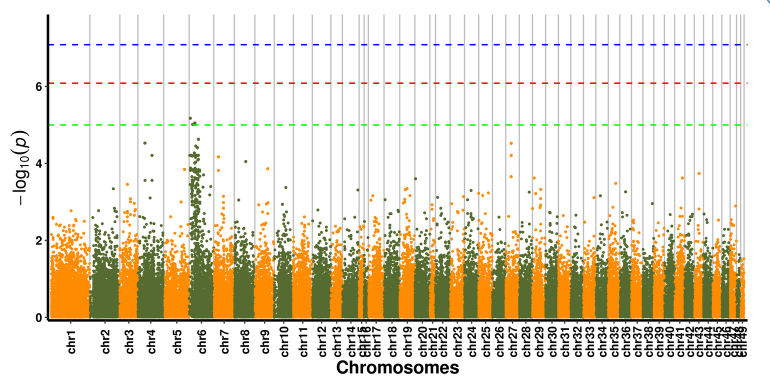
Ka nānā ʻana o ka hui SNP-trait - Manhattan plot
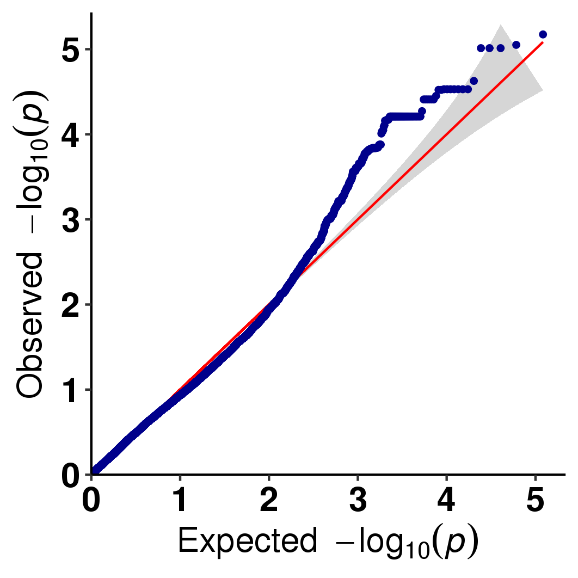
Ka nānā 'ana i ka hui SNP-trait - QQ plot
E ʻimi i nā holomua i hoʻomaʻamaʻa ʻia e nā lawelawe de GWAS o BMKGene ma o ka hōʻiliʻili ʻana o nā puke.
Lv, L. et al. (2023) 'Insight in the genetic basis of ammonia tolerance in razor clam Sinonovacula constricta by genome-wide association study',ʻAiʻa, 569, p. 739351. doi: 10.1016/J.AQUACULTURE.2023.739351.
Li, X. et al. (2022) 'O nā loiloi multi-omics o 398 foxtail millet accessions hōʻike i nā wahi genomic e pili ana i ka home, nā ʻano metabolite, a me nā hopena anti-inflammatory',Mea Molekula, 15(8), pp. 1367–1383. doi: 10.1016/j.molp.2022.07.003.
Li, J. et al. (2022) 'Genome-Wide Association Mapping of Hulless Barely Phenotypes in Drought Environment',Nā palena i ka ʻepekema mea kanu, 13, p. 924892. doi: 10.3389/FPLS.2022.924892/BIBTEX.
Zhao, X. et al. (2021) 'GmST1, ka mea i hoʻopili i ka sulfotransferase, hāʻawi i ke kū'ē i ka soybean mosaic virus strains G2 a me G3',Mea kanu, Cell & Environment, 44(8), pp. 2777–2792. doi: 10.1111/PCE.14066.